- A: a/b three-layered sandwiches
- X: Flavodoxin-like
- H: Class I glutamine amidotransferase-like
- T: Rossmann-like domain in dehydroquinate synthase-like enzymes
- F: DHQ_synthase
| UID: | 000359486 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1sg6A2 |
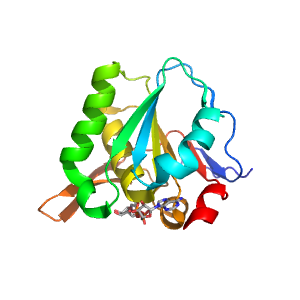
| Range: | B:4-163 |
| Ligands: | NAD |
| PDB: | 3clh |
| PDB Description: | 3-DEHYDROQUINATE SYNTHASE |
| UniProt: | P56081 |
| Species: | Helicobacter pylori |