- A: a+b two layers
- X: FAD-linked reductases, C-terminal domain-like
- H: FAD-linked reductases-C
- T: FAD-linked reductases-C
- F: GMC_oxred_C
| UID: | 000359723 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1ju2A1 |
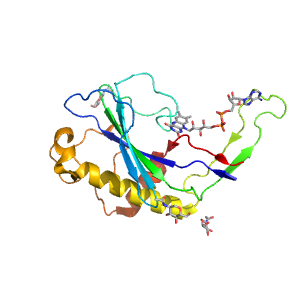
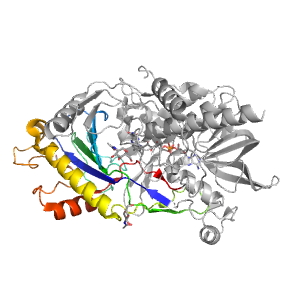
| Range: | A:294-463 |
| Ligands: | FAD NAG |
| PDB: | 3gdp |
| PDB Description: | R-OXYNITRILE LYASE ISOENZYME 1 |
| UniProt: | Q945K2 |
| Species: | Prunus dulcis |