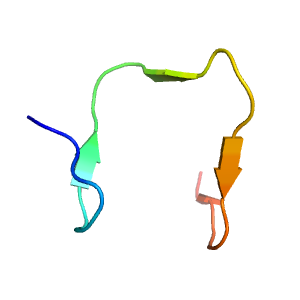
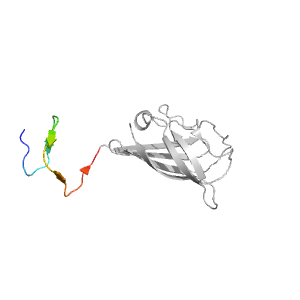
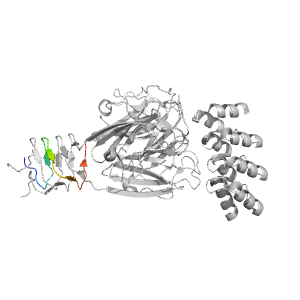
- A: beta duplicates or obligate multimers
- X: Phage tail fiber protein trimerization domain
- H: Phage tail fiber protein trimerization domain
- T: Phage tail fiber protein trimerization domain
- F: BppL_N
| UID: | 001150335 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2f0cA2 |
| Range: | B:34-62 |
| PDB: | 3hg0 |
| PDB Description: | BASEPLATE PROTEIN |
| UniProt: | Q9G096 |
| Species: | Lactococcus phage TP901-1 |