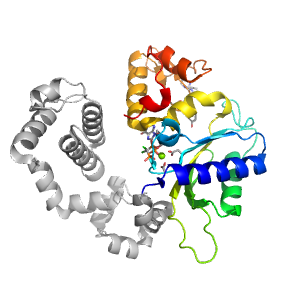
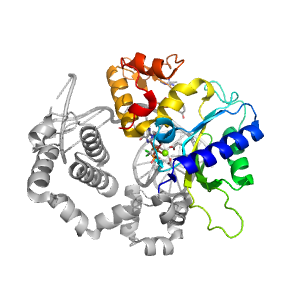
- A: a+b three layers
- X: Nucleotidyltransferase-like
- H: Nucleotidyltransferase
- T: Nucleotidyltransferase
- F: DNA_pol_B_thumb,DNA_pol_B_palm
| UID: | 000392280 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2fmpA3 |
| Range: | A:149-335 |
| Ligands: | CL G2C MG |
| PDB: | 3jpn |
| PDB Description: | DNA POLYMERASE BETA |
| UniProt: | P06746 |
| Hsap BLAST neighbor: | P06746 |
| Species: | Homo sapiens |