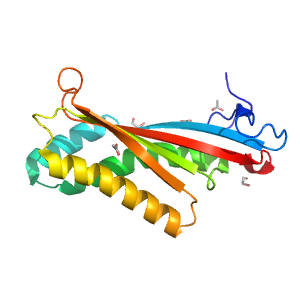
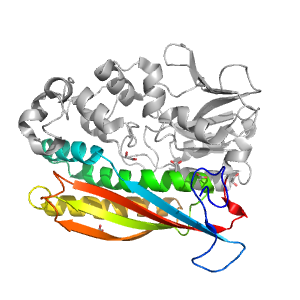
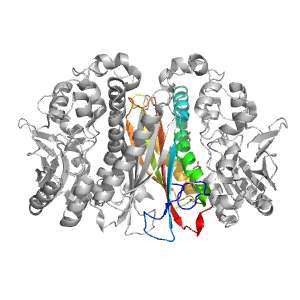
- A: a+b three layers
- X: Bacillus chorismate mutase-like
- H: PurM N-terminal domain-like
- T: PurM N-terminal domain-like
- F: AIRS
| UID: | 001150148 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1cliA2 |
| Range: | A:27-174 |
| Ligands: | ACT EDO |
| PDB: | 3kiz |
| PDB Description: | Phosphoribosylformylglycinamidine cyclo-ligase |
| UniProt: | Q11YU7 |
| Species: |