- A: a/b three-layered sandwiches
- X: Flavodoxin-like
- H: Class I glutamine amidotransferase-like
- T: CheY-like
- F: B12-binding
| UID: | 001337999 |
|---|---|
| Type: | Automatic domain |
| Parent: | e3bulA3 |
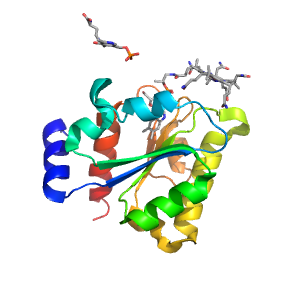
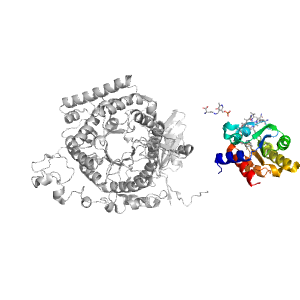
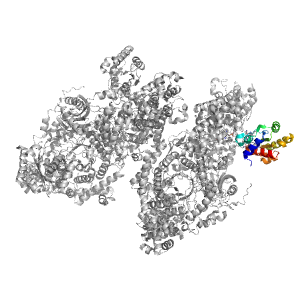
| Range: | B:591-740 |
| Ligands: | B12 Z98 |
| PDB: | 3kox |
| PDB Description: | D-ornithine aminomutase E component |
| UniProt: | E3PY95 |
| Species: | [Clostridium] sticklandii |