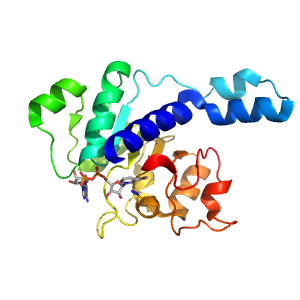
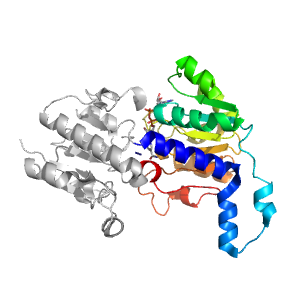
- A: a/b three-layered sandwiches
- X: Rossmann-like
- H: Rossmann-related
- T: NAD(P)-binding Rossmann-fold domains
- F: 2-Hacid_dh_C
| UID: | 000393044 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2nacA1 |
| Range: | K:146-335 |
| Ligands: | AZI NAD |
| PDB: | 3n7u |
| PDB Description: | Formate dehydrogenase |
| UniProt: | Q9S7E4 |
| Hsap BLAST neighbor: | O43175 |
| Species: | Arabidopsis thaliana |