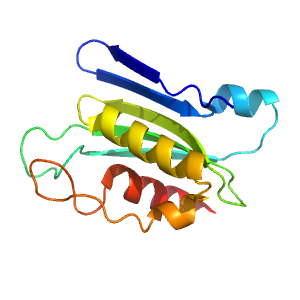
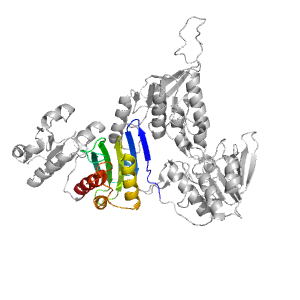
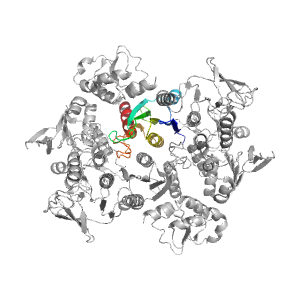
- A: a+b two layers
- X: FAD-linked reductases, C-terminal domain-like
- H: FAD/NAD-linked reduatases, dimerisation (C-terminal) domain
- T: FAD/NAD-linked reduatases, dimerisation (C-terminal) domain
- F: Pyr_redox_dim
| UID: | 001770212 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1nhpA1 |
| Range: | A:344-468 |
| Ligands: | COA FAD |
| PDB: | 3nta |
| PDB Description: | FAD-DEPENDENT PYRIDINE NUCLEOTIDE-DISULPHIDE OXIDOREDUCTASE |
| UniProt: | A3QAV3 |
| Species: |