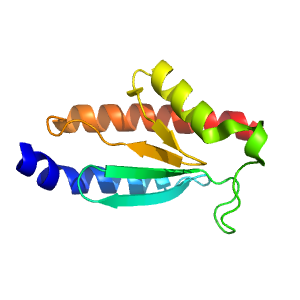
- A: a+b two layers
- X: C-terminal domain in some PLP-dependent transferases
- H: C-terminal domain in some PLP-dependent transferases
- T: C-terminal domain in some PLP-dependent transferases
- F: Aminotran_3
| UID: | 001505965 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1zodA2 |
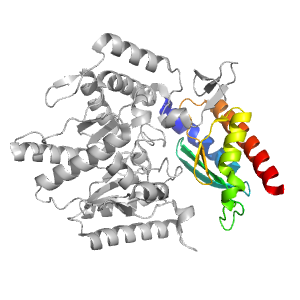
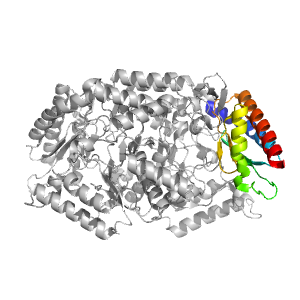
| Range: | A:345-454 |
| PDB: | 3nui |
| PDB Description: | Pyruvate transaminase |
| UniProt: | F2XBU9 |
| Species: | Vibrio fluvialis |