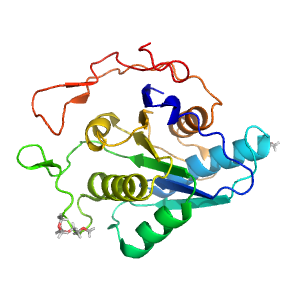
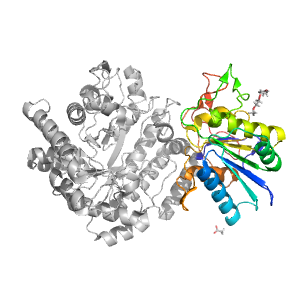
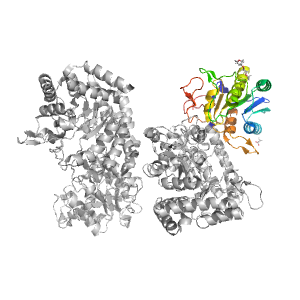
- A: a/b three-layered sandwiches
- X: Flavodoxin-like
- H: Beta-D-glucan exohydrolase, C-terminal domain
- T: Beta-D-glucan exohydrolase, C-terminal domain
- F: Glyco_hydro_3_C
| UID: | 000140073 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1x38A2 |
| Range: | A:435-642 |
| Ligands: | ACT PG4 |
| PDB: | 3nvd |
| PDB Description: | UNCHARACTERIZED LIPOPROTEIN YBBD |
| UniProt: | P40406 |
| Species: | Bacillus subtilis |