- A: beta sandwiches
- X: Immunoglobulin-like beta-sandwich
- H: Immunoglobulin-related
- T: Cu,Zn superoxide dismutase-like
- F: Sod_Cu
| UID: | 001168135 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2c9vA1 |
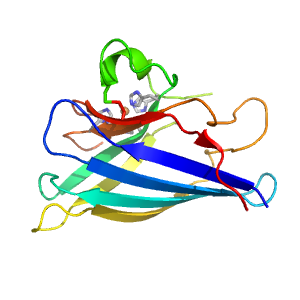
| Range: | C:2-154 |
| Ligands: | CU |
| PDB: | 3s0p |
| PDB Description: | SUPEROXIDE DISMUTASE [CU-ZN], CHLOROPLASTIC |
| UniProt: | P14831 |
| Hsap BLAST neighbor: | P00441 |
| Species: | Solanum lycopersicum |