- A: a+b three layers
- X: Thioredoxin-like
- H: Thioredoxin-like
- T: Thioredoxin-like
- F: AhpC-TSA,1-cysPrx_C
| UID: | 001197416 |
|---|---|
| Type: | Automatic domain |
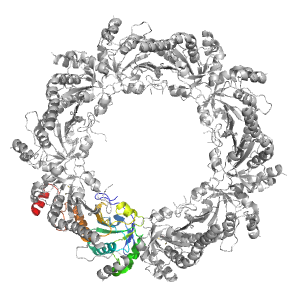
| Parent: | e1qmvA1 |
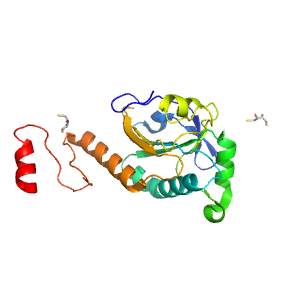
| Range: | D:1-194 |
| Ligands: | DTU DTV |
| PDB: | 3sbc |
| PDB Description: | Peroxiredoxin TSA1 |
| UniProt: | P34760 |
| Hsap BLAST neighbor: | P32119 |
| Species: | Saccharomyces cerevisiae |