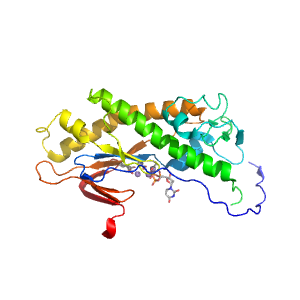
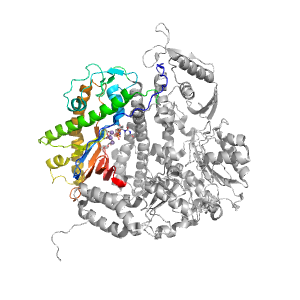
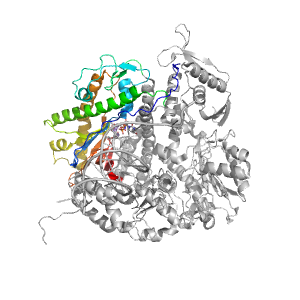
- A: a+b two layers
- X: Alpha-beta plaits
- H: Adenylyl and guanylyl cyclase catalytic domain-like
- T: Adenylyl and guanylyl cyclase catalytic domain-like
- F: DNA_pol_B
| UID: | 001256843 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1tgoA4 |
| Range: | A:376-468,A:575-732 |
| Ligands: | DUP MN |
| PDB: | 3sjj |
| PDB Description: | DNA POLYMERASE |
| UniProt: | Q38087 |
| Species: | Enterobacteria phage RB69 |