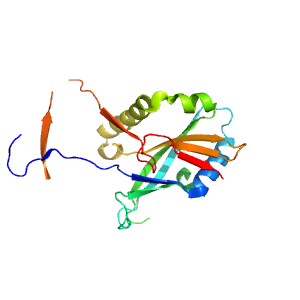
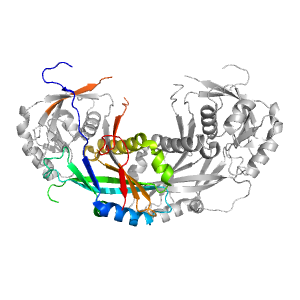
- A: a/b three-layered sandwiches
- X: Atg7 N-terminal domain-like
- H: N-terminal domain in E1 enzyme Atg7
- T: N-terminal domain in E1 enzyme Atg7
- F: ATG7_N
| UID: | 000433838 |
|---|---|
| Type: | Automatic domain |
| Parent: | e3vx7A1 |
| Range: | A:-1-139,A:250-288 |
| PDB: | 3t7h |
| PDB Description: | UBIQUITIN-LIKE MODIFIER-ACTIVATING ENZYME ATG7 |
| UniProt: | P38862 |
| Hsap BLAST neighbor: | O95352 |
| Species: | Saccharomyces cerevisiae |