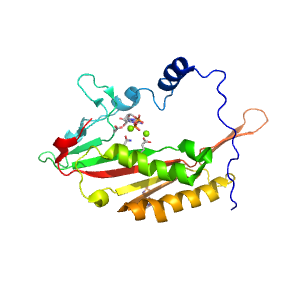
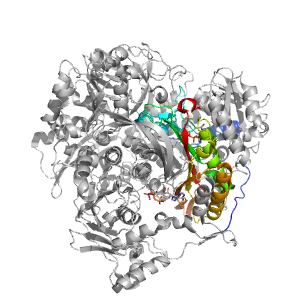
- A: a+b three layers
- X: Bacillus chorismate mutase-like
- H: PurM N-terminal domain-like
- T: PurM N-terminal domain-like
- F: FGAR-AT_PurM_N-like
| UID: | 001150152 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1t3tA6 |
| Range: | A:616-816 |
| Ligands: | ADP ANP MG SO4 |
| PDB: | 3umm |
| PDB Description: | PHOSPHORIBOSYLFORMYLGLYCINAMIDINE SYNTHASE |
| UniProt: | P74881 |
| Hsap BLAST neighbor: | O15067 |
| Species: |