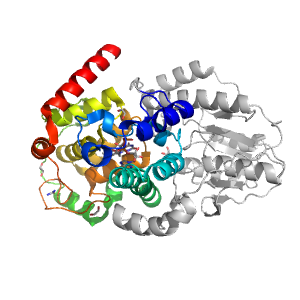
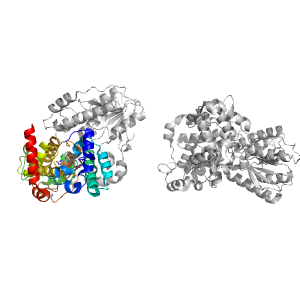
- A: alpha complex topology
- X: Cryptochrome/photolyase FAD-binding domain-related
- H: Cryptochrome/photolyase FAD-binding domain-related
- T: Cryptochrome/photolyase FAD-binding domain
- F: Unmapped domains
| UID: | 001164975 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2j07A1 |
| Range: | A:242-490 |
| Ligands: | EDO FAD URE |
| PDB: | 3umv |
| PDB Description: | DEOXYRIBODIPYRIMIDINE PHOTO-LYASE |
| UniProt: | Q6F6A2 |
| Species: |