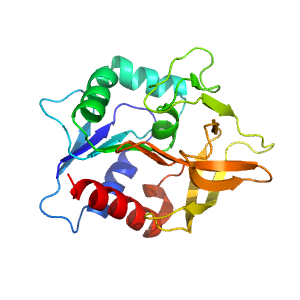
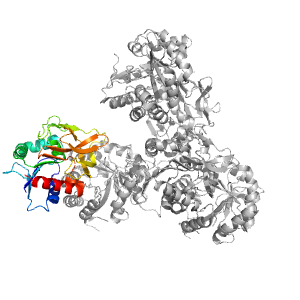
- A: a/b three-layered sandwiches
- X: Flavodoxin-like
- H: Class I glutamine amidotransferase-like
- T: Class I glutamine amidotransferase-like
- F: GATase
| UID: | 000988962 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1gpmA3 |
| Range: | F:1-201 |
| PDB: | 3zr4 |
| PDB Description: | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISH |
| UniProt: | Q9X0C8 |
| Species: | Thermotoga maritima |