- A: a/b three-layered sandwiches
- X: PLP-dependent transferases
- H: PLP-dependent transferases
- T: PLP-dependent transferases
- F: Aminotran_3
| UID: | 001759332 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1zodA3 |
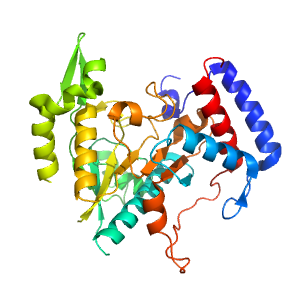
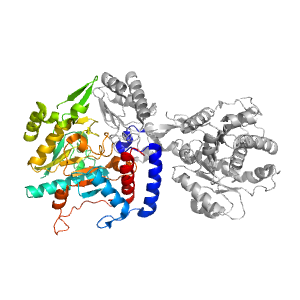
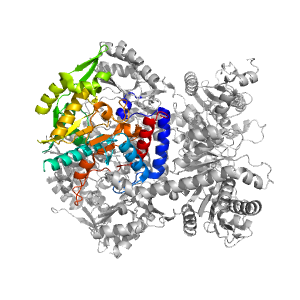
| Range: | A:364-703 |
| Ligands: | PLP |
| PDB: | 4a0f |
| PDB Description: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE |
| UniProt: | B0F481 |
| Hsap BLAST neighbor: | P04181 |
| Species: | Arabidopsis thaliana |