- A: few secondary structure elements
- X: Rubredoxin-like
- H: Rubredoxin-related
- T: Rubredoxin-related
- F: Unmapped domains
| UID: | 001170981 |
|---|---|
| Type: | Automatic domain |
| Parent: | e2b4yA1 |
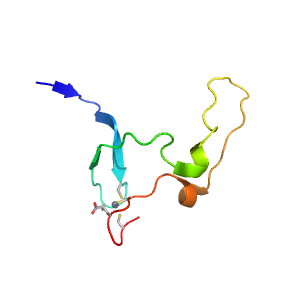
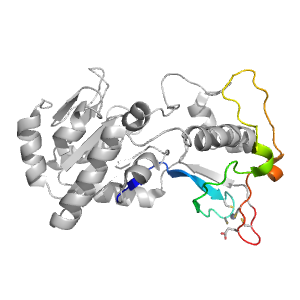
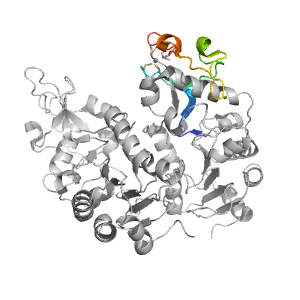
| Range: | A:153-213 |
| Ligands: | ZN |
| PDB: | 4hda |
| PDB Description: | NAD-dependent protein deacylase sirtuin-5, mitochondrial |
| UniProt: | Q9NXA8 |
| Hsap BLAST neighbor: | Q9NXA8 |
| Species: | Homo sapiens |