| UID: | 001569083 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1nzaA1 |
| Range: | X:1-103 |
| PDB: | 4zk7 |
| PDB Description: | Divalent-cation tolerance protein CutA |
| UniProt: | Q7SIA8 |
| Hsap BLAST neighbor: | O60888 |
| Species: |
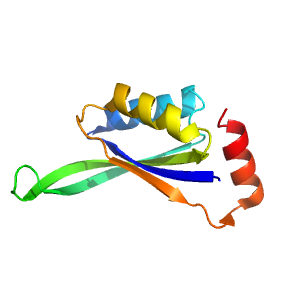
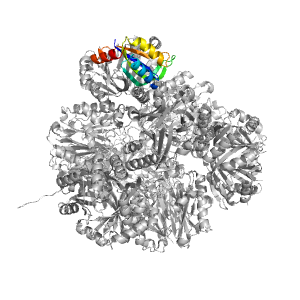
Structure of domain e4zk7X1
Domains in the same chain:
| e4zk7X1 | X:1-103 | Alpha-beta plaits | GlnB-like | GlnB-like |
Domains in the same PDB: