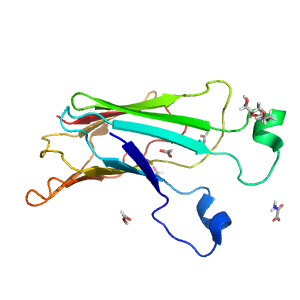
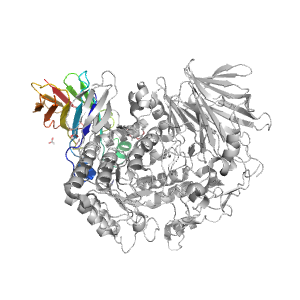
- A: beta sandwiches
- X: Putative glucosidase YicI, C-terminal domain
- H: Putative glucosidase YicI, C-terminal domain
- T: Putative glucosidase YicI, C-terminal domain
- F: Unmapped domains
| UID: | 001868446 |
|---|---|
| Type: | Automatic domain |
| Parent: | e3l4uA4 |
| Range: | A:829-966 |
| Ligands: | FMT OXM P6G |
| PDB: | 5hjr |
| PDB Description: | Neutral alpha-glucosidase AB |
| UniProt: | Q8BHN3 |
| Hsap BLAST neighbor: | Q14697 |
| Species: | Mus musculus |