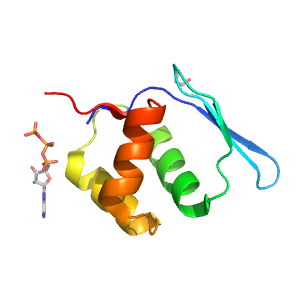
| UID: | 002514592 |
|---|---|
| Type: | Automatic domain |
| Parent: | e6gos11 |
| Range: | 2:13-90 |
| Ligands: | ATP EDO |
| PDB: | 6grg |
| PDB Description: | Microcin B17-processing protein McbB |
| UniProt: | P23184 |
| Species: |
Structure of domain e6grg22
Domains in the same chain:
| e6grg22 | 2:13-90 | HTH | HTH | winged |
| e6grg21 | 2:91-294 | Rossmann-like | Rossmann-related | Activating enzymes of the ubiquitin-like proteins |
Domains in the same PDB: