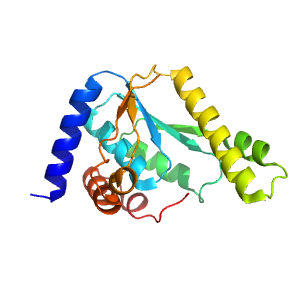
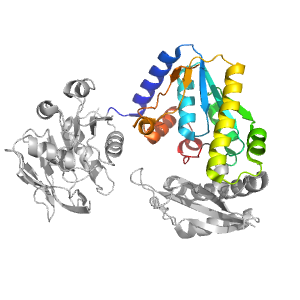
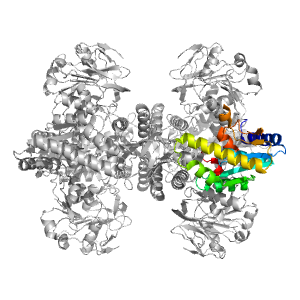
- A: a/b three-layered sandwiches
- X: HUP domain-like
- H: HUP domains
- T: HUP domains
- F: Unmapped domains
| UID: | 000330161 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1kqpA1 |
| Range: | B:189-322,B:347-382 |
| PDB: | 2ywb |
| PDB Description: | GMP synthase [glutamine-hydrolyzing] |
| UniProt: | Q5SI28 |
| Hsap BLAST neighbor: | P49915 |
| Species: |