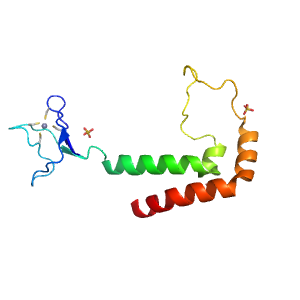
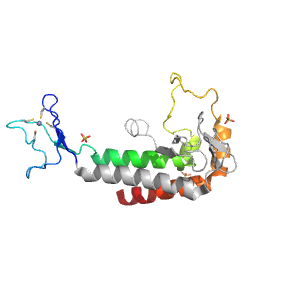
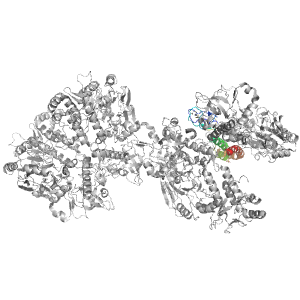
- A: few secondary structure elements
- X: Rubredoxin-like
- H: Zinc finger domain of DNA polymerase-alpha
- T: Zinc finger domain of DNA polymerase-alpha
- F: zf-DNA_Pol
| UID: | 000364895 |
|---|---|
| Type: | Automatic domain |
| Parent: | e3floB2 |
| Range: | F:1345-1452 |
| Ligands: | SO4 ZN |
| PDB: | 3flo |
| PDB Description: | DNA polymerase alpha catalytic subunit A |
| UniProt: | P13382 |
| Species: | Saccharomyces cerevisiae |