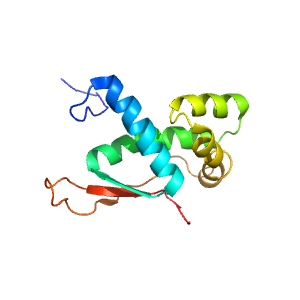
- A: a+b two layers
- X: Enolase-N/ribosomal protein
- H: Prokaryotic ribosomal protein L17
- T: Prokaryotic ribosomal protein L17
- F: Ribosomal_L17
| UID: | 001125021 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1gd8A1 |
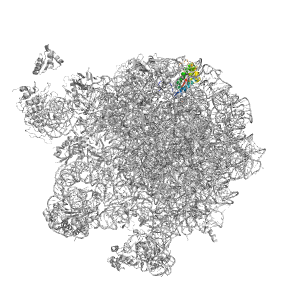
| Range: | Q:1-120 |
| PDB: | 3j5w |
| PDB Description: | |
| Hsap BLAST neighbor: | Q9NRX2 |
| Species: |