- A: a/b three-layered sandwiches
- X: Flavodoxin-like
- H: Class I glutamine amidotransferase-like
- T: Periplasmic binding protein-like I
- F: Peripla_BP_1
| UID: | 001547493 |
|---|---|
| Type: | Automatic domain |
| Parent: | e8abpA3 |
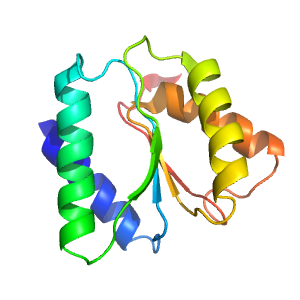
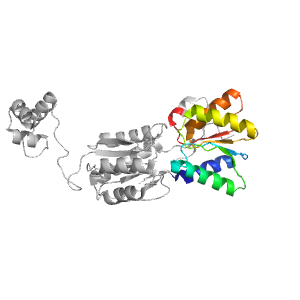
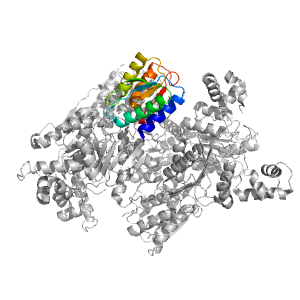
| Range: | C:166-285 |
| PDB: | 3kjx |
| PDB Description: | Transcriptional regulator, LacI family |
| UniProt: | Q5LM80 |
| Species: | Ruegeria pomeroyi |