- A: alpha bundles
- X: CO dehydrogenase ISP C-domain like
- H: CO dehydrogenase ISP C-domain like
- T: CO dehydrogenase ISP C-domain like
- F: Fer2_2
| UID: | 000410385 |
|---|---|
| Type: | Automatic domain |
| Parent: | e1v97A1 |
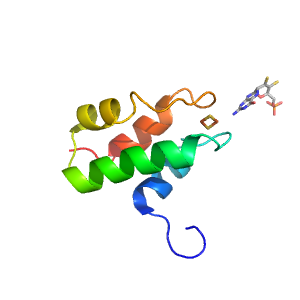
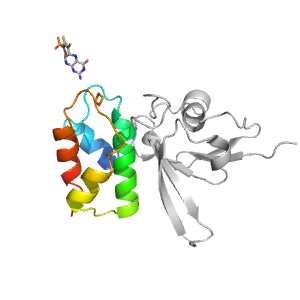
| Range: | A:93-165 |
| Ligands: | FES MTE |
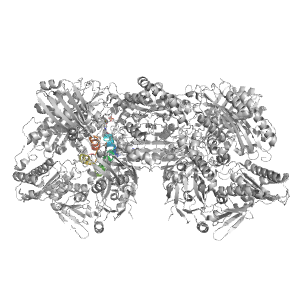
| PDB: | 3nvv |
| PDB Description: | XANTHINE DEHYDROGENASE/OXIDASE |
| UniProt: | P80457 |
| Hsap BLAST neighbor: | P47989 |
| Species: | Bos taurus |