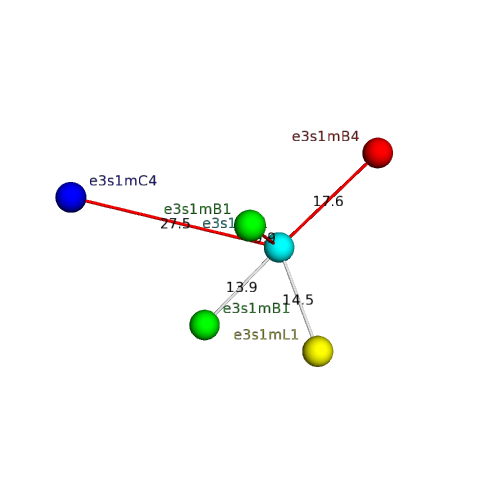
| Domain ID | Symmetry operator | H-group | Visualization |
|---|
| e3s1mB1 |
L:x,y,z->B:X,Y,Z |
alpha/beta-Hammerhead/Barrel-sandwich hybrid |
Interaction
Interface
Pymol
|
| e3s1mB6 |
L:x,y,z->B:X,Y,Z |
N-terminal domain in beta subunit of DNA dependent RNA-polymerase |
Interaction
Interface
Pymol
|
| e3s1mC4 |
L:x,y,z->C:X,Y,Z |
Insert subdomain of RNA polymerase alpha subunit |
Interaction
Interface
Pymol
|
| e3s1mL1 |
L:x,y,z->L:-X,Y,-Z |
Rubredoxin-related |
Interaction
Interface
Pymol
|
| e3s1mL1 |
L:-X,Y,-Z->L:x,y,z |
Rubredoxin-related |
Interaction
Interface
Pymol
|
| e3s1mB1 |
L:x,y,z->B:-X,Y,-Z |
alpha/beta-Hammerhead/Barrel-sandwich hybrid |
Interaction
Interface
Pymol
|