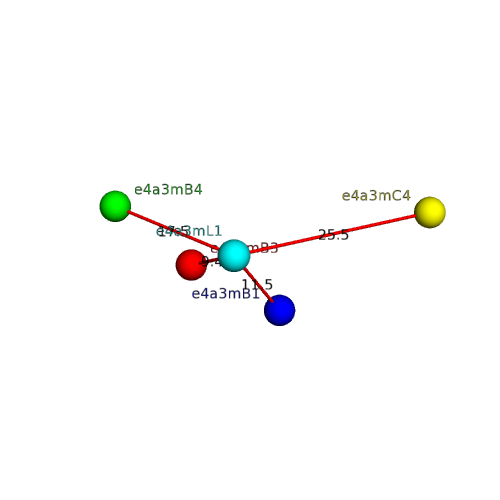
| e4a3mA3 |
A:3-336 |
N-terminal domain in RNA-polymerase beta-prime subunit |
N-terminal domain in RNA-polymerase beta-prime subunit |
N-terminal domain in RNA-polymerase beta-prime subunit |
| e4a3mA6 |
A:337-374,A:436-519 |
cradle loop barrel |
RIFT-related |
double psi |
| e4a3mA7 |
A:375-435 |
MoeA-I/Ornithine decarboxylase-C/Reverse ferredoxin-like domain in RNA-polymerase |
MoeA-I/Ornithine decarboxylase-C/Reverse ferredoxin-like domain in RNA-polymerase |
MoeA-I/Ornithine decarboxylase-C/Reverse ferredoxin-like domain in RNA-polymerase |
| e4a3mA8 |
A:520-662 |
first helical domain in RNA-polymerase beta-prime subunit |
first helical domain in RNA-polymerase beta-prime subunit |
first helical domain in RNA-polymerase beta-prime subunit |
| e4a3mA2 |
A:663-871 |
second helical domain in RNA-polymerase beta-prime subunit |
second helical domain in RNA-polymerase beta-prime subunit |
second helical domain in RNA-polymerase beta-prime subunit |
| e4a3mA9 |
A:872-1059 |
helical domain in yeast RNA-polymerases |
third helical domain in yeast RNA-polymerase II beta-prime subunit |
third helical domain in yeast RNA-polymerase II beta-prime subunit |
| e4a3mA4 |
A:1060-1102,A:1301-1455 |
central helical domain in RNA-polymerase beta-prime subunit |
central helical domain in RNA-polymerase beta-prime subunit |
central helical domain in RNA-polymerase beta-prime subunit |
| e4a3mA5 |
A:1103-1131,A:1243-1300 |
ferredoxin-like domain in yeast RNA-polymerase beta-prime subunit |
ferredoxin-like domain in yeast RNA-polymerase beta-prime subunit |
ferredoxin-like domain in yeast RNA-polymerase beta-prime subunit |
| e4a3mA1 |
A:1132-1242 |
permuted ferredoxin-like domain in yeast RNA-polymerase beta-prime subunit |
permuted ferredoxin-like domain in yeast RNA-polymerase beta-prime subunit |
permuted ferredoxin-like domain in yeast RNA-polymerase beta-prime subunit |
| e4a3mB6 |
B:20-70,B:90-134,B:164-218,B:405-541 |
N-terminal domain in beta subunit of DNA dependent RNA-polymerase |
N-terminal domain in beta subunit of DNA dependent RNA-polymerase |
N-terminal domain in beta subunit of DNA dependent RNA-polymerase |
| e4a3mB3 |
B:219-404 |
insertion domain in beta subunit of DNA dependent RNA-polymerase |
insertion domain in beta subunit of DNA dependent RNA-polymerase |
insertion domain in beta subunit of DNA dependent RNA-polymerase |
| e4a3mB2 |
B:542-630 |
inserted a+b domain in yeast RNA polymerase beta subunit |
inserted a+b domain in yeast RNA polymerase beta subunit |
inserted a+b domain in yeast RNA polymerase beta subunit |
| e4a3mB7 |
B:631-813 |
barrel domain in beta subunit of DNA dependent RNA-polymerase |
barrel domain in beta subunit of DNA dependent RNA-polymerase |
barrel domain in beta subunit of DNA dependent RNA-polymerase |
| e4a3mB5 |
B:814-850,B:975-1097 |
cradle loop barrel |
RIFT-related |
double psi |
| e4a3mB4 |
B:851-974 |
alpha/beta-Hammerhead/Barrel-sandwich hybrid |
alpha/beta-Hammerhead/Barrel-sandwich hybrid |
Single hybrid motif |
| e4a3mB1 |
B:1098-1224 |
C-terminal domain in beta subunit of DNA dependent RNA-polymerase |
C-terminal domain in beta subunit of DNA dependent RNA-polymerase |
C-terminal domain in beta subunit of DNA dependent RNA-polymerase |
| e4a3mC3 |
C:3-41,C:173-268 |
DCoH-like |
RBP11-like subunits of RNA polymerase |
RBP11-like subunits of RNA polymerase |
| e4a3mC4 |
C:42-172 |
Ribosomal protein TL5/L25 C-terminal domain-like |
Insert subdomain of RNA polymerase alpha subunit |
Insert subdomain of RNA polymerase alpha subunit |
| e4a3mD1 |
D:3-71,D:131-221 |
HhH/H2TH |
SAM/DNA-glycosylase |
SAM domain-like |
| e4a3mE2 |
E:2-142 |
Restriction endonuclease-like |
Eukaryotic RPB5 N-terminal domain |
Eukaryotic RPB5 N-terminal domain |
| e4a3mE1 |
E:143-215 |
RPB5-like RNA polymerase subunit |
RPB5-like RNA polymerase subunit |
RPB5-like RNA polymerase subunit |
| e4a3mF1 |
F:72-155 |
RPB6/omega subunit-like |
RPB6/omega subunit-like |
RPB6 |
| e4a3mG4 |
G:1-68 |
Dodecin subunit-like |
Flavin-binding protein dodecin-like |
Flavin-binding protein dodecin-like |
| e4a3mG3 |
G:69-171 |
OB-fold |
Nucleic acid-binding proteins |
Nucleic acid-binding proteins |
| e4a3mH1 |
H:2-146 |
OB-fold |
Nucleic acid-binding proteins |
Nucleic acid-binding proteins |
| e4a3mI1 |
I:2-49 |
Rubredoxin-like |
Rubredoxin-related |
Rubredoxin-related |
| e4a3mI2 |
I:50-120 |
Rubredoxin-like |
Rubredoxin-related |
Rubredoxin-related |
| e4a3mJ1 |
J:1-65 |
HTH |
RPB10 |
RPB10 |
| e4a3mK1 |
K:1-115 |
DCoH-like |
RBP11-like subunits of RNA polymerase |
RBP11-like subunits of RNA polymerase |
| e4a3mL1 |
L:25-70 |
Rubredoxin-like |
Rubredoxin-related |
Rubredoxin-related |