Research
"Nothing in biology makes sense
except in the light of evolution."
T. Dobzhansky (1900-1975)
We develop and use theoretical methods to study proteins
We work at the interface of biology, computer science, mathematics and physics. Our group specializes in computational biology of proteins and combines sequence and structure analysis with evolutionary considerations to facilitate discoveries of biological significance. Two major directions are pursued:
|
This duality, i.e. combination of methods development with biological applications is beneficial for both directions, as we frequently find that existing approaches do not give satisfactory answers to specific biological questions; and availability of experts biologists in our group to validate the results in the process, stimulates methods development.
• Introduction •
What is the most important unsolved problem in computational biology of proteins? Apparently, protein energetics. 1) Protein folding problem (i.e. prediction of spatial structure from sequence); 2) precise modeling of interactions between proteins and of proteins with other molecules; 3) quantitative understanding of enzyme catalysis — are all various incarnations of the same challenge scientists were not able to overcome. Despite significant achievements, the exact solution from the physics perspective is still far from reach.
Bioinformatics approaches offer practically useful shortcuts to these problems. Deduction of protein properties (i.e. 3D structure or function) by homology to proteins with known properties has been most successful. For this method to reach its full potential, the following steps should be perfected. We need to: 1) find homologous proteins, i.e. do a database search; 2) compare them to the protein of interest, e.g. make an alignment; 3) decide on the boundaries of property transfer by similarity, i.e. at what level of similarity a property is shared between homologs, and thus can be deduced for a homolog without available experimental information from experimentally characterized homolog. Most projects in the lab deal with these questions.
Homology and evolution are the central themes of our research. By homology we mean similarity caused by common ancestry, not any kind of similarity. Proteins are homologous if they originated from a common ancestor. Similarity caused by other reasons, e.g. structural constraints on 3D packing, is termed analogy. When similarity is weak, it is not trivial to distinguish between the two scenarios leading to similarities between proteins: homology or analogy, and we are working on this problem.
• Main Research Directions •
The long-term objective of our research is to classify available sequence-structure data on proteins into a biologically relevant, hierarchical system analogous to the one currently used in zoology and botany, and to provide computational tools to maintain and update this classification. Since sequence and structural similarities usually imply functional similarity, such classification is of indispensable value for biologists to aid in experimental design. Combination of various approaches is best for addressing complex problems. Thus the questions we pursue are quite diverse and can be summarized as follows.
|
|
Our publications give more specific ideas about research directions. We also are involved in many collaborations to study individual protein families, predict properties of proteins, help interpret the effects of clinical mutations and assist in data analysis–driven experimental design.
• Research Highlights •
A. Method development
During the last few years, we have been working on improvement of computational methods to assess protein similarity. We mainly concentrated on two levels of similarity between proteins: sequence and structure. We also directly addressed structure prediction as an important application of homology inference.
A.1. Sequence similarity detection methods
Two main steps constitute sequence analysis. First, homologues are found and separated from unrelated proteins; second, these homologues are aligned. We suggested strategies to improve both steps.
A.1.1. COMPASS – comparison of multiple sequence alignments
Compass is a profile-profile aligner based on a solid probabilistic basis.
A.1.2. Multiple Sequence Alignment
In the process of a tedious search for a tool to provide fast and accurate alignments, we developed this program that up until now demonstrated the best alignment accuracy. PCMA combines COMPASS scoring with the consistency strategy of T-Coffee
B.1.3. Sequence design assists homology searches
Sequence diversity in alignments is essential for
profile-based methods. Profile improvement can be achieved by enriching
alignments with more sequences sharing a common fold. These sequences can
either come from sequencing projects, or be designed in silico.
Structure-based design explores small but important covariation between
positions, which is
being ignored by profile methods. We extended the ideas of Koehl&Levitt to
cover sequence-based searches. The design procedure may involve sequences that
are expected to fold into a known structure36,37, or
ancestral sequences reconstructed from a multiple sequence alignment38.
Implementing both strategies, we demonstrated that they assist homology
inference.
To capitalize on these preliminary experiments, we will
combine evolutionary and structural design strategies to produce in silico
sequences for many protein families, and will carry out sequence searches in
the new expanded space.
B.2.
Structure similarity detection methods
None of the numerous methods proposed for structure
similarity search39-42 achieves
what we consider the most fundamental task, i.e. finding structural motifs that
satisfy the general definition of a protein fold: globular units with the same
secondary structure, topology and architecture irrespective of subtle
differences in packing between elements. Such a program should be key in
protein structure-mining. The closest available utility, TOPS43 falls
short, since it checks topology only. While developing a structural motif
search algorithm, we were surprised to find that a method providing broad, yet
accurate secondary structure delineation44,45 was lacking
as well.
B.2.1. PAL8E – delineation of secondary structural
elements
We delineate secondary structural elements using Ca
coordinates in order to generate input for the fold search46. The
elements we define (helices and strands) are linear, so they can be
approximated by vectors; and cover about 85% of residues in the structure, so
they can provide maximum representation in an element-based search. Our program
is predictive by nature, i.e. it does not evaluate the actual existence of
hydrogen bonds as DSSP44, but rather
checks if the general geometry is sufficiently close to provide H-bond presence
in some conditions. As such, PAL8E is robust to coordinate errors up to 1.5Å
RMSD and is more suitable as the first step to finding remote structural
similarities.
B.2.2. ProSMoS – Protein Structure Motif Search
Programs that assess structural similarity compare two
structures to each other and define common regions39-42. Structural
classification experts look for a particular structural motif instead10,47. Most
programs base similarity scores on superposition and closeness of either
coordinates or contacts. Experts pay more attention to the main chain
orientation and general match of secondary structural elements. We developed a
program that emulates an expert48. Starting
from a structure, we use PAL8E46 to
delineate secondary structural elements. A matrix of element interactions (parallel
or antiparallel) and handedness of connections is constructed. All structures
are reduced to matrices that contain just enough information to define a fold,
so the definition is very general and large deviations in coordinates are
tolerated. A user supplies a matrix for a motif, and ProSMoS lists all
structures that exactly match this motif. Application of ProSMoS to several
tasks demonstrated its usefulness49-51.
We have yet to 1) develop a good scoring function; 2)
implement an algorithm that finds partial matches to the motif, and 3) develop
an algorithm to produce a residue-to-residue alignment based on detected motif
similarity.
B.3.
Combination of methods for homology detection: SCOPmap
Likely the most difficult task is to identify whether
proteins with remotely similar structures share a common ancestor9. Done
largely by expert analysis, such work is very time-consuming10,47. To tackle
it computationally, we designed a strategy that checks sequence and structure
similarity statistics using various existing programs, combines their scores,
and attributes a protein structure to a previously defined evolutionary
classification52. Our
automatic method assigns about 95% of proteins to SCOP leaving the rest to
expert analysis. Instances of incorrect assignments near domain boundaries need
more work.
B.4.
Structure modeling methods
After establishing a homology link, the structure of one
protein can be modeled using the other one as a template53. We tried
to improve scoring functions for several specific problems in homology
modeling.
B.4.1. Side-chain modeling
Placement of given side-chains on a fixed backbone
(=packing prediction) is a necessary step towards a realistic protein model54. Careful
refinement of weights for “energy” terms and usage of contact
surface/overlapped volume allowed us to obtain a better side-chain rotamer
predictor55, which has
been used by many other researchers, e.g.56-58.
B.4.2. Sequence design
Finding optimal side-chains for a fixed backbone
(=protein sequence prediction) is a natural extension of the side-chain
modeling problem59. Parameters
refined on a set of high resolution structures correlated well with
experimental data on mutant T4 lysozymes stability37. We used
the designed sequences to assist in remote homology detection.
We plan to 1) incorporate restricted backbone moves in
the design algorithm; 2) as a collaboration with Dr. Zhang, to determine the
X-ray structure of a protein with a new fold that we constructed de novo
from short fragments of the backbone and designed a sequence for with our
approach.
B.4.3. Local structure prediction
Fragment-based structure prediction methods60 explore
“horizontal” (local sequence) information in protein sequences, in contrast to
sequence alignment methods that use “vertical” (positional) information
assuming independence between positions61. To improve
homology detection and sequence alignment construction using local conformation
predictions, we developed a local fragment predictor28.
We plan to add statistical significance estimation of
local profile hits and to incorporate it into alignment algorithms.
B.5.
Methods to evaluate the quality of structural models
Our group was invited to judge predictions in Critical
Assessment of Structure Prediction competition (CASP5)62. Evaluating
thousands of structural models, we found63 that a
consensus method combining many scores generated by various structure
similarity scoring schemes correlates best with manual assessment of models
that has been a standard in former CASPs. Approaches we developed have been
successfully used by different assessors in last year’s CASP6.
C.
Application to biological problems
As our main scientific interest is to find novel remote
homology links between proteins, we applied various computational methods
enhanced with the manual expert analysis to explore sequence and structure
databases.
C.1.
Large-scale evolutionary classification of proteins
The ultimate goal of our research is to classify all
protein domains according to their evolutionary history. Moving towards this
goal, we selected several protein groups for in-depth analysis. These groups
can be structural, i.e. all proteins of a particular fold; functional, i.e. all
proteins of a particular function; or genome-based, i.e. all proteins in a
particular genome.
C.1.1. Fold-based
Small protein domains (20-60 residues) are notoriously
difficult to analyze due to unreliable similarity statistics caused by
insufficient numbers of residues64. In an
attempt to bring order to one of the small protein groups, we classified all
zinc finger structures.
C.1.1.1. Zinc fingers
We define zinc fingers (ZF) as small domains where zinc
contributes to the protein structural core. We classify all ZF structures into
8 fold groups based on structural similarity around the zinc-binding site65. Each
fold-group encompasses 1 to 11 evolutionary families. Prior to our work, there
has been no targeted attempt to comprehensively classify all ZFs. However,
rudiments of such classification existed within general structure databases10,47.
We are currently extending our experience with ZFs over
another very large group of small proteins: disulphide-rich domains.
C.1.1.2. Thioredoxin fold
For larger domains with the core formed by secondary
structural elements, classification could start with ProSMoS search48, as we have
done for thioredoxin fold, an a/b
domain of 6 secondary structural elements50. Our
definition of fold allows circular permutations, i.e. geometric transformations
that link existing N- and C-termini of the chain and introduce new termini at
another point. Since permutations regularly happen in protein evolution66, their
inclusion in the fold definition ensures completeness of coverage. We unify
about 1000 protein structures from 5 different SCOP folds10, and divide
them into 11 evolutionary families. For many of these proteins, structural
similarity to thioredoxin remained unreported.
We are working on classification of RNAse H and Rossmann
folds, and will plan to cover other wide-spread folds, such as Greek-key
immunoglobulin-like, ferredoxin-like and a-folded leaf.
C.1.2. Function-based: kinases
Bringing together all proteins that share significant
functional similarities and looking at the whole group from evolutionary
perspective is informative for understanding both function and evolution67,68. In
addition, such work results in many challenging structure predictions. Our
study of kinases defined as enzymes transferring terminal P-group from ATP to
another molecule is included among the five references69,70. Although
protein kinases are frequently analyzed and classified71, no
comprehensive classification of all kinases, including many families of
metabolic small molecule kinases, existed.
We are extending our experience with kinases on several
evolutionary diverse protein functional classes, such as proteases,
phosphatases and acyltransferases.
C.1.3. Genome-based: human Y chromosome
Computational genomics necessitates annotation of
proteins encoded in complete genomes72. Meticulous
expert-driven study of proteins from a single organism is capable of finding
interesting functional annotations and structure predictions that are missed in
large-scale automatic analyses73. We applied
our experience to human Y-chromosome74 (see one of
the five publications).
We are planning to undertake diligent domain analysis of
all human proteins with the goal of evolutionary classification and structural
prediction, crucial for our understanding of human biology.
C.2.
Studies of individual protein families
This aspect of our work is arguably closest to the bench
and richest in immediate biological applications. Taking an individual protein
and a few of its relatives, we embark upon solving a problem that other
researchers pursue at the bench, namely, we try to find out what that protein
does and how it functions. Many of such cases result in unexpected structure
predictions.
C.2.1. Structure predictions
During the last few years, our studies of two dozen
families resulted in non-trivial and yet confident structure predictions75-90 Many of our
published predictions remain unverified by experimental structures. However,
more structures become available every year. Some of these correspond to
proteins for which we have published predictions. On occasion, structural
biologists discuss our predictions in their publications describing
experimental structures91. Several
such articles are included in the Appendix to this report. So far, not a single
fold prediction reported by us has been found incorrect. Naturally, careful
analysis of alignments reveals that some details of our models are not always
accurate. The most interesting deviation from the model was found by crystal
structures of GyrA and ParC domains92,93 that were
predicted to form six-bladed b–propellers82. Crystal
structures revealed that both domains are indeed comprised of six blades with 4
b–strands
each. However, each blade is involved in a swap of 2 b–strands
with the adjacent blade. This swap could not be deduced by us.
We plan to continue our work on non-trivial structure
predictions of interesting domains.
C.2.2. Homology predictions
The most important aspect of our research is remote
homology prediction, i.e. deduction that proteins A and B share a common
ancestor despite low similarity between them. Although such statement cannot be
verified by direct experimentation, it predicts the possibility of existence of
a protein C, such that both links between A and C, and between C and B are
strong and clear. Thus, homology between A and B can be verified by
transitivity. Protein C may be currently unknown, but frequently its sequence
or structure eventually becomes available. The best argument for remote
homology prediction is made when multiple evidence (sequence, structure, function)
is considered94-103. Despite
remoteness of homology, it could be helpful for the functional understanding of
a protein. One such example, MH1 domain of SMAD, is discussed in an included
publication100.
Our runs of SCOPmap program52 on weekly
updates of PDB frequently produce possible, but not fully justified
assignments. We will be inspecting them to find new non-trivial homology links
that we could support by manual analysis.
C.3.
Fold changes in protein evolution
Analyzing distantly related proteins, at times we see
structural changes large enough to warrant placing homologues in different
folds. We were pioneers in recognizing this universal trend104,105. We try to
understand how protein structures, usually considered to be rigid, change in evolution106,107. One
particular aspect of the problem is distinction between homologues and
analogues, and finding examples of analogues108-110.
Our goal for the next several years is to comprehensively
catalogue all instances of homologues with significant structural differences,
and to understand mechanisms used most commonly by evolution to change protein
structures.
• More info about significant projects •
Fold Change in Evolution of Proteins
From the early days of protein structural biology, researches have been surprised by the resistance of protein spatial structures to evolutionary changes. This remarkable structural robustness combined with the limited number of available 3D structures has lead to a view that the abstract protein structure space is discrete, can be divided into a number of folds, and protein evolution mostly proceeds within the framework of the same fold.
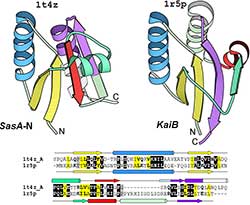
Today, with the rapidly increasing number of protein structures, arguably, the majority of protein structural patterns have been experimentally determined and a new view of structural continuity of folding patterns is starting to emerge. Many examples of proteins with statistically significant sequence similarity, but substantial structural differences, have been documented. Such phenomenon demonstrates the evolutionary bridges between structurally different proteins and profoundly influences our understanding of protein structure evolution. On one hand, the notion that protein structures are evolutionarily plastic and changeable has important applications in protein design and opens new frontiers in engineering proteins that possess desired functional properties, such as a possibility to create proteins with condition-dependent folds. On the other hand, the existence of proteins with similar sequences but different structures hinders homology modeling methods; therefore our ability to detect such cases from sequence is crucial. To study the mechanisms and paths of protein fold change in evolution, we undertook comprehensive comparative analysis of protein sequences and structures, and catalogued the instances of potentially homologous proteins with significant structural differences. Our work revealed that, although such instances are not very common, they are universally observed among proteins of all structural classes, and involve substantial structural changes and rearrangements that may be explained by both small sequence changes, such as point mutations, and large sequence rearrangements, such as non-homologous recombination.
« lessMultiple Sequence Alignment (MSA)
The accurate multiple sequence alignment (MSA) of protein sequences is essential for a wide range of applications from structure modeling to prediction of functional sites [1, 2]. Today, almost every study of a protein starts from MSA construction. MSA of a few homologs reveals patterns of sequence conservation, pointing researchers to important regions in protein sequences. Despite this overwhelming need, MSAs are not very accurate when sequence identity drops below 25%.
Thus, construction of high quality MSA is considered to be the second (after folding) biggest unsolved problem in computational biology. Over the years, our group developed several MSA applications [3-5]. The most recent one is the program PROMALS, which uses secondary structure predictions and a powerful scoring function [4, 6]. PROMALS is the best MSA program according to our test and a recently published review by a pioneer in MSA methods [2]. For the sequences with low similarity, PROMALS is about three times more accurate than the well-known software ClustalW [7]. However, for very distant sequences with identity around 15%, only ~40% of amino acids are aligned correctly by PROMALS. Thus further improvement is needed.
« less