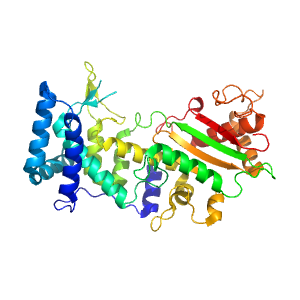
- A: a+b two layers
- X: Alpha-beta plaits
- H: Adenylyl and guanylyl cyclase catalytic domain-like
- T: Adenylyl and guanylyl cyclase catalytic domain-like
- F: RRM_4
| UID: | 001309339 |
|---|---|
| Type: | Automatic domain |
| Parent: | e3zefE1 |
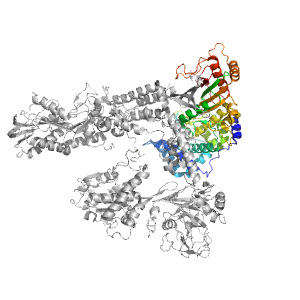
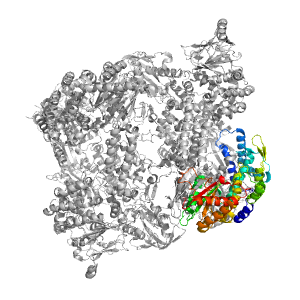
| Range: | B:883-1255 |
| PDB: | 3zef |
| PDB Description: | PRE-MRNA-SPLICING FACTOR 8 |
| UniProt: | P33334 |
| Hsap BLAST neighbor: | Q6P2Q9 |
| Species: | Saccharomyces cerevisiae |