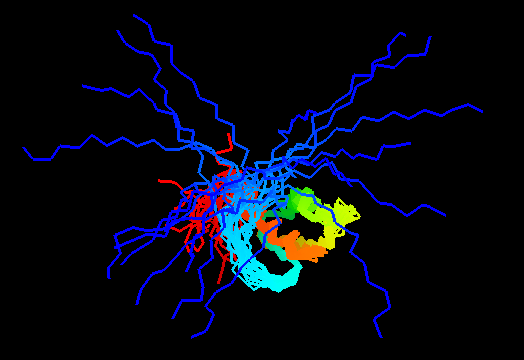
{F,S}loppy NMR Structures
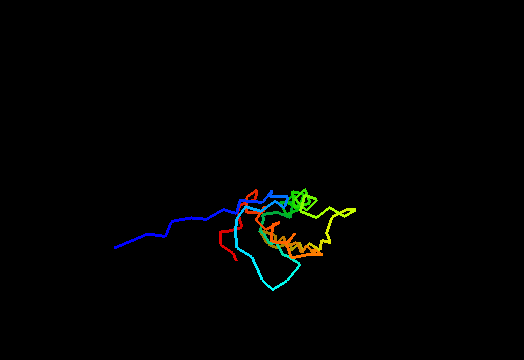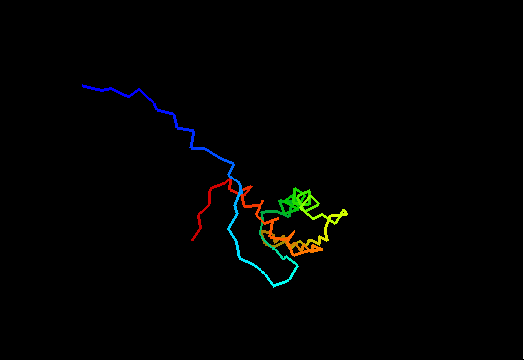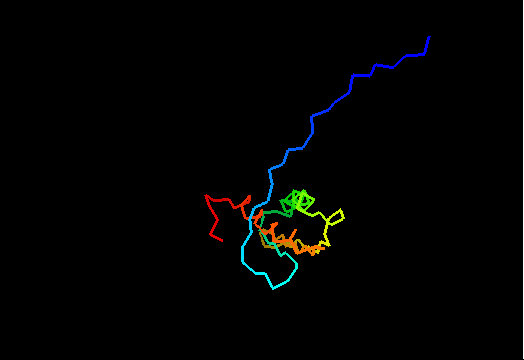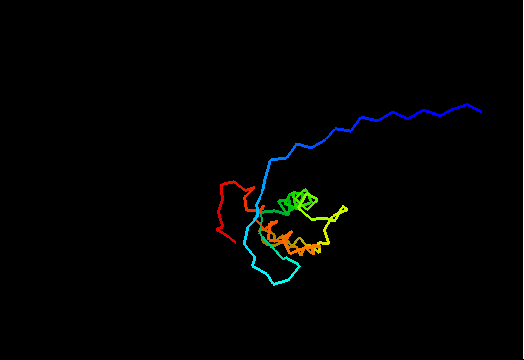
T0437 (2k3i): protein yiiS from Shigella flexneri
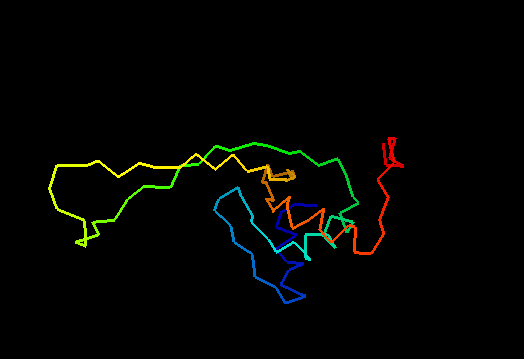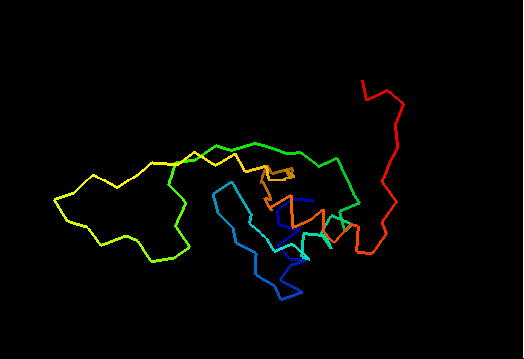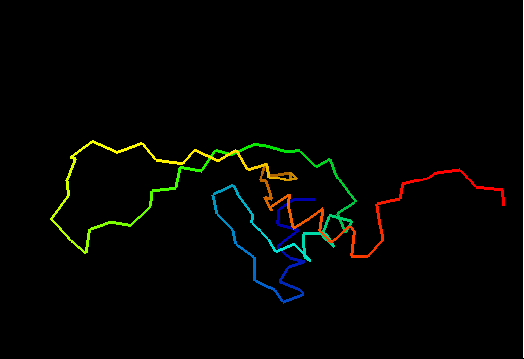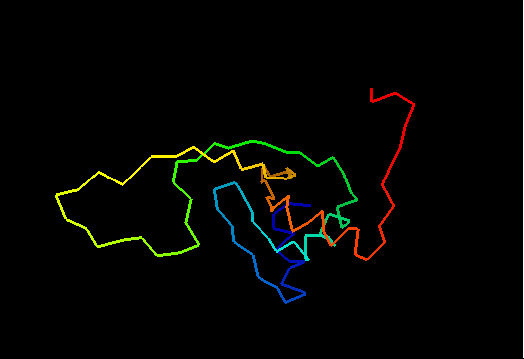
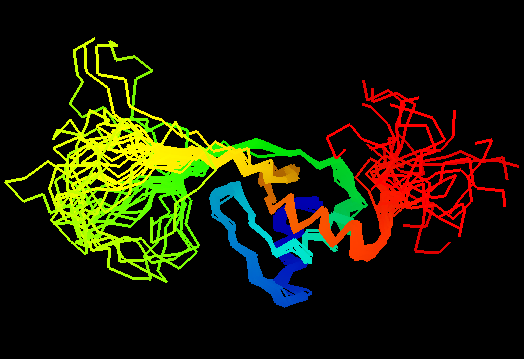
T0460 (2k4n): protein PF0246 from Pyrococcus furiosus
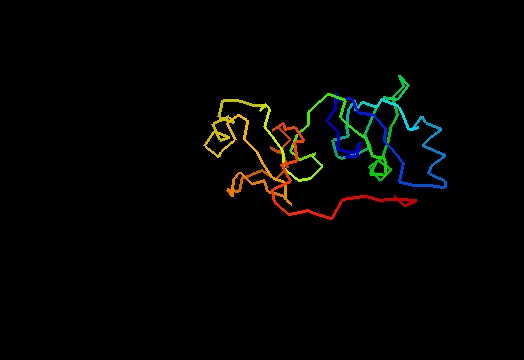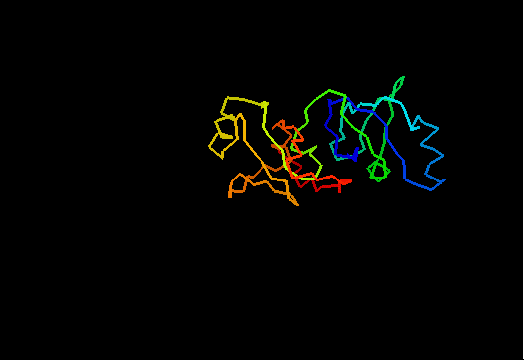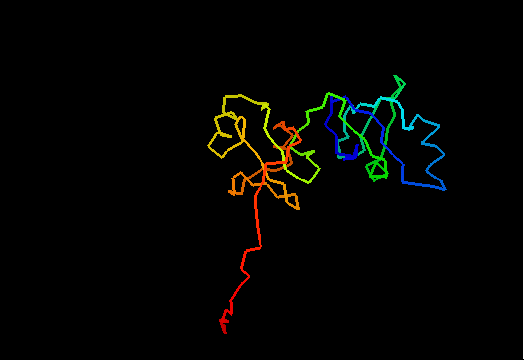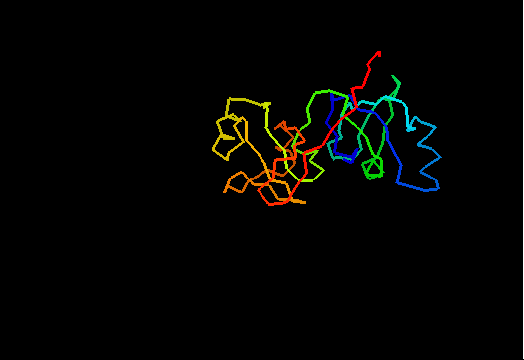
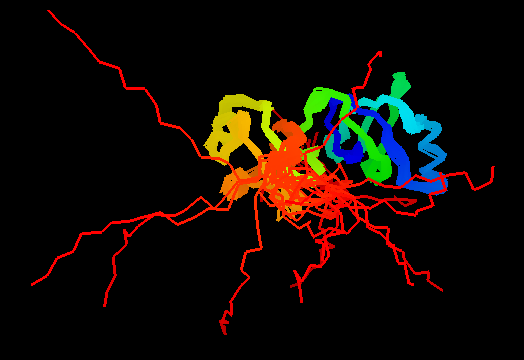
T0462 (2k5i): iron(ii) transport protein from Clostridium thermocellum
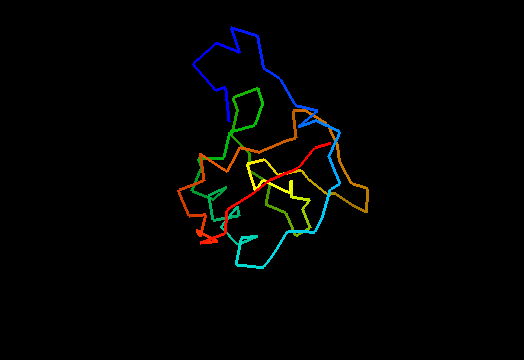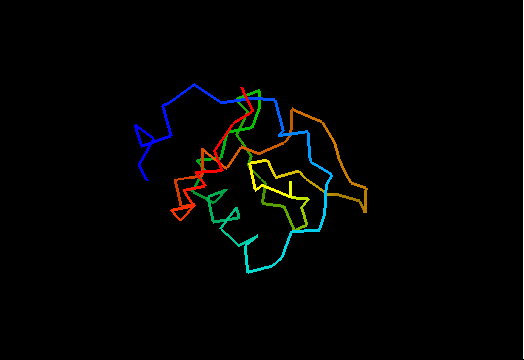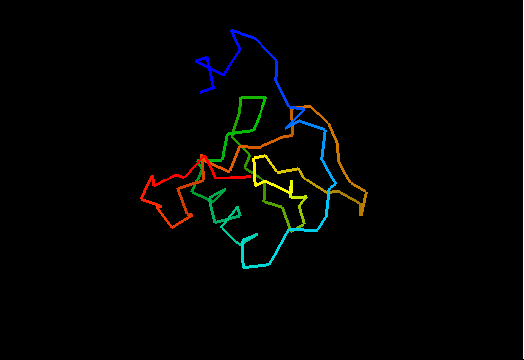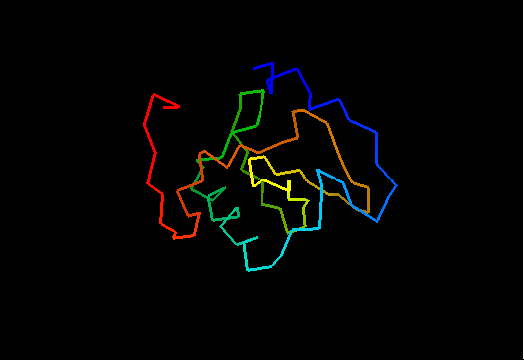
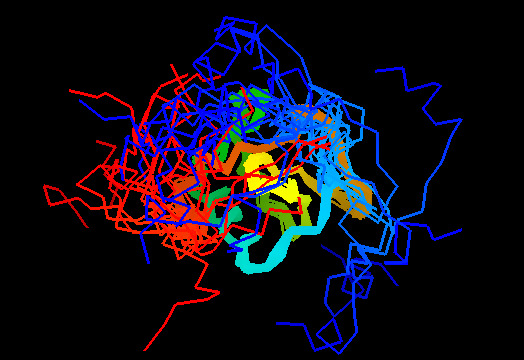
T0464 (2k5r): XF2673 from Xylella fastidiosa
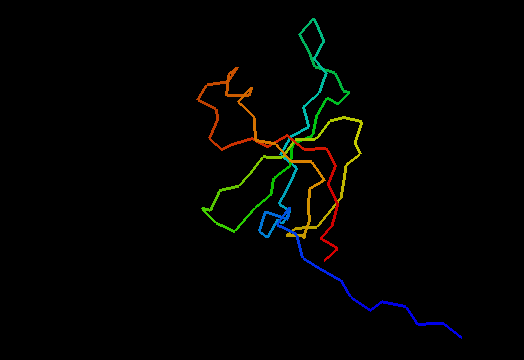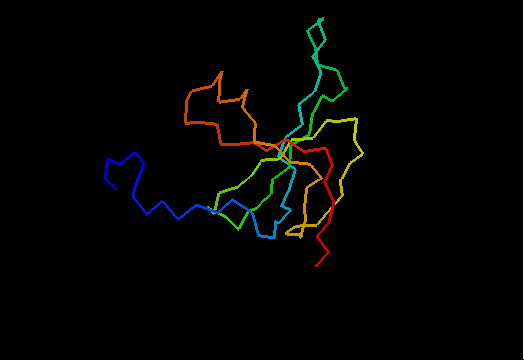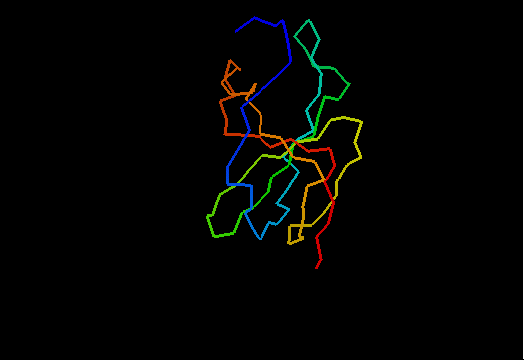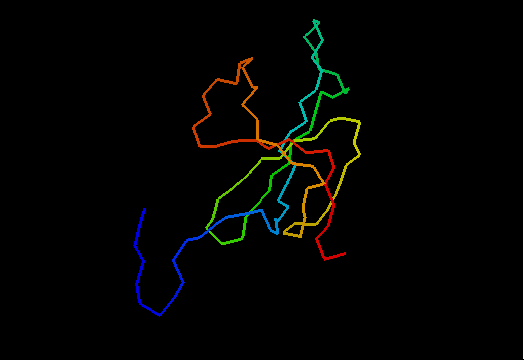
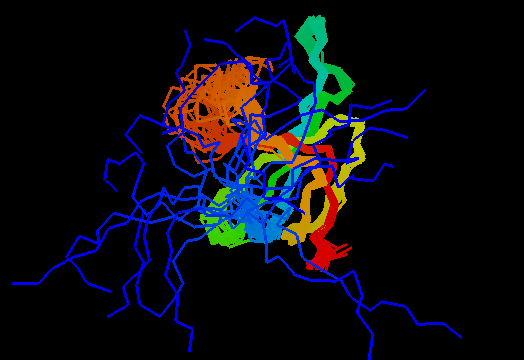
T0466 (2k5d): SAG0934 from Streptococcus agalactiae
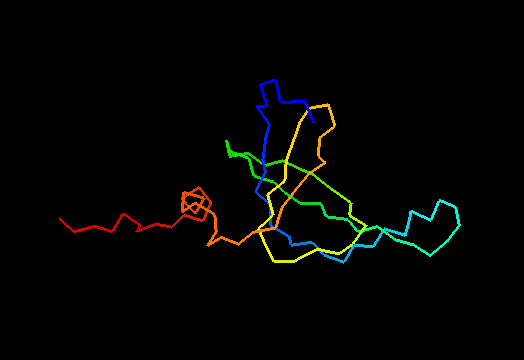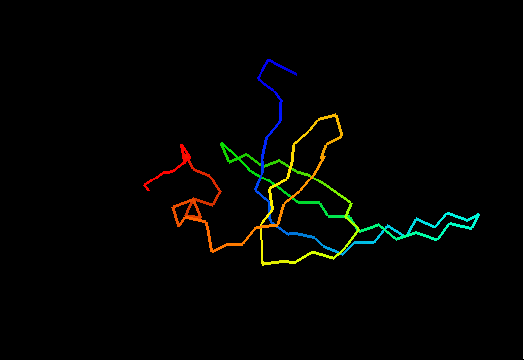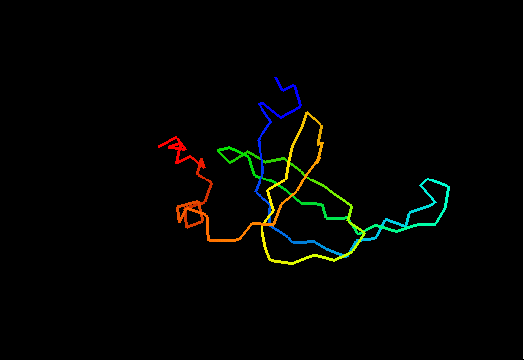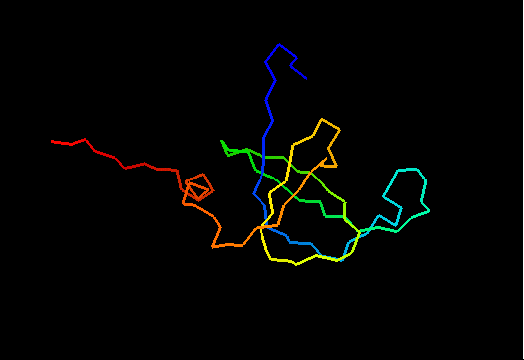
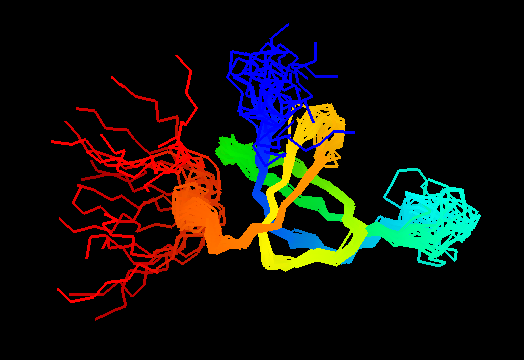
T0467 (2k5q): membrane associated protein from Bacillus cereus
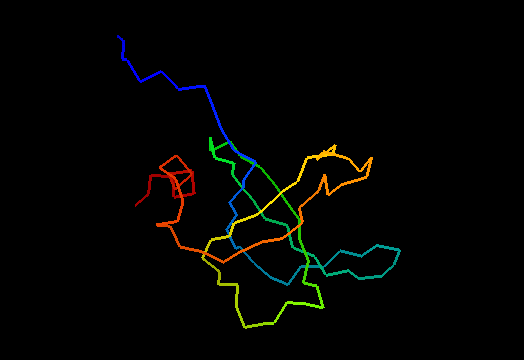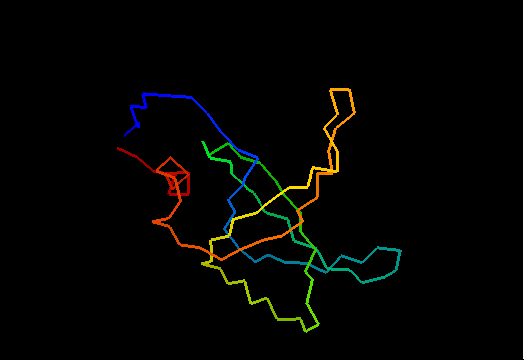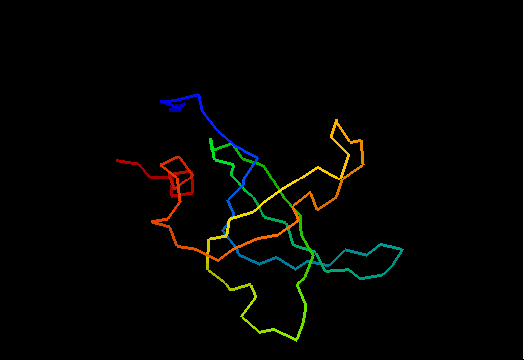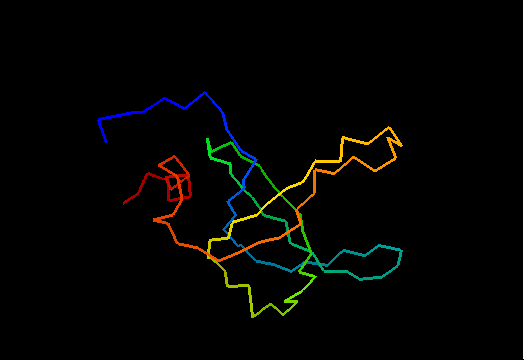
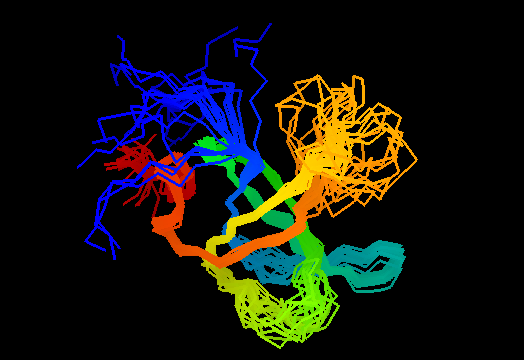
T0468 (2k5w): putative lipoprotein from Bacillus cereus
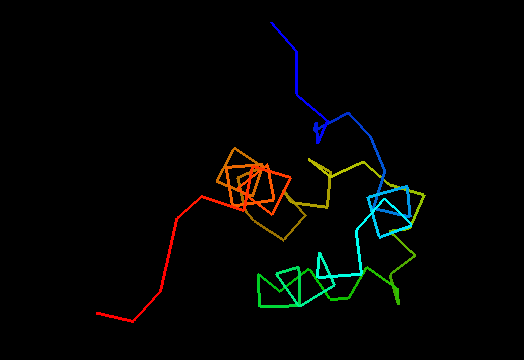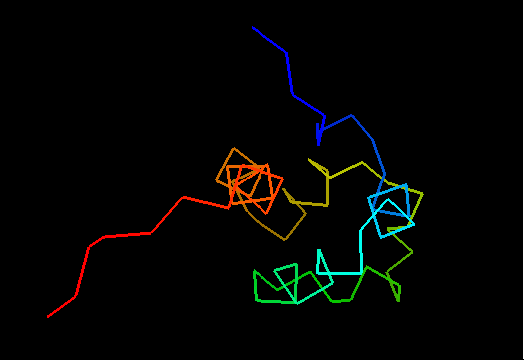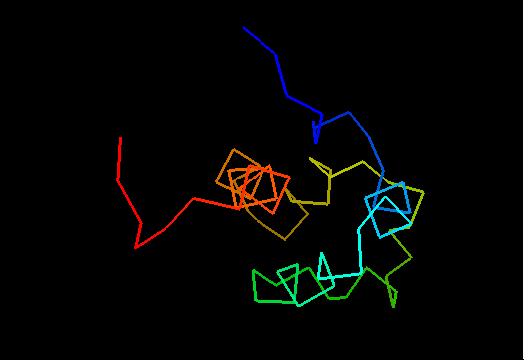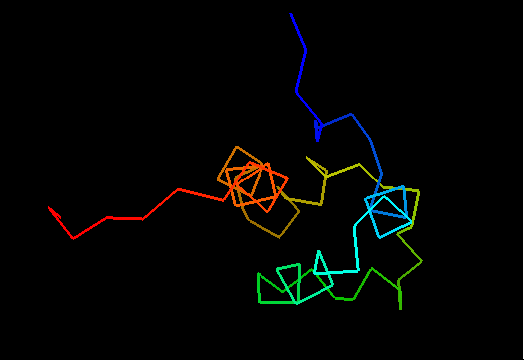
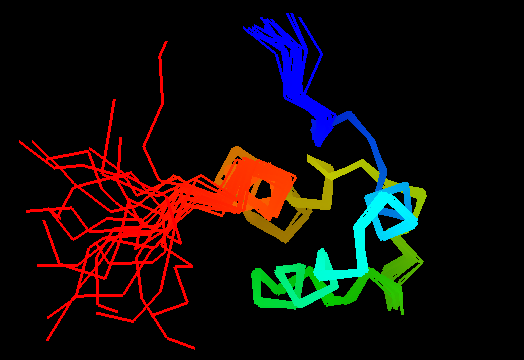
T0469 (2k5e): protein GSU1278 from Methanococcus jannaschii
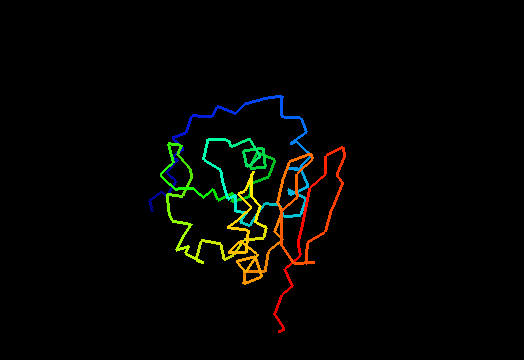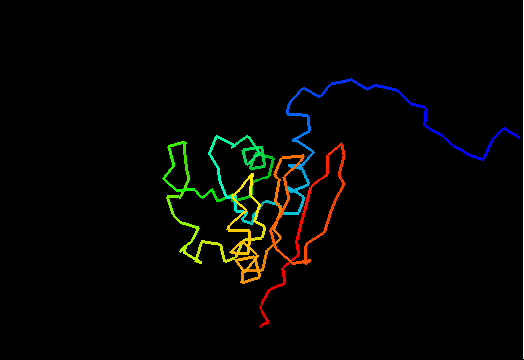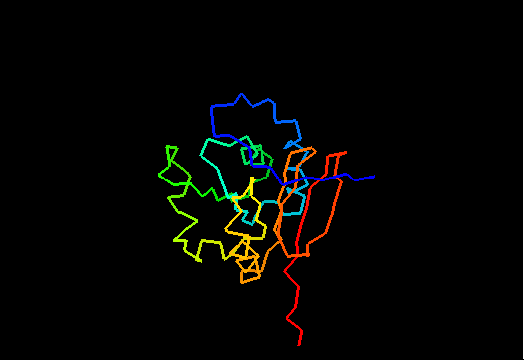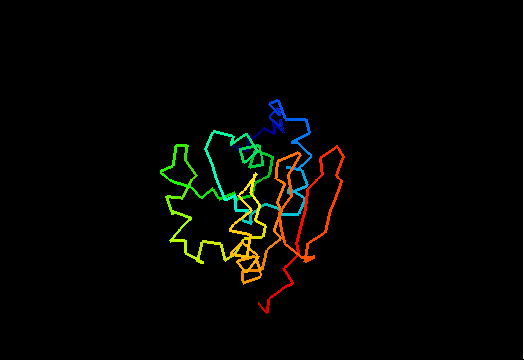
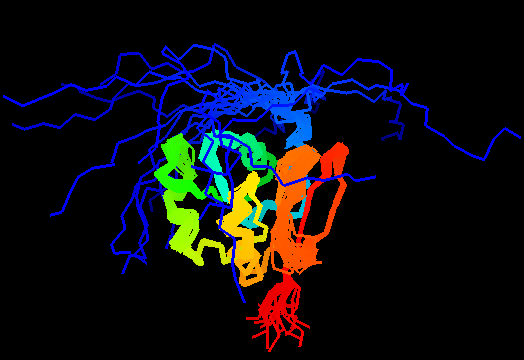
T0471 (2k4m): methyltransferase MTH_1000 from Methanobacterium thermoautotrophicum
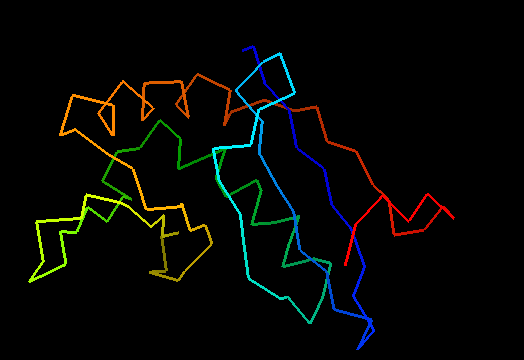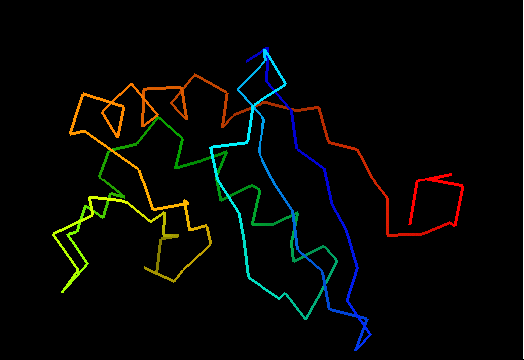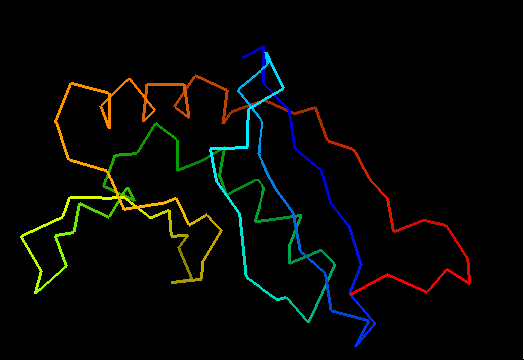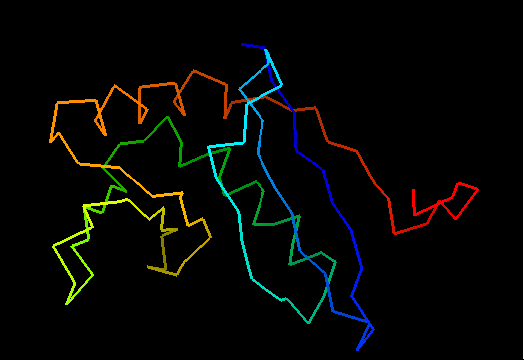
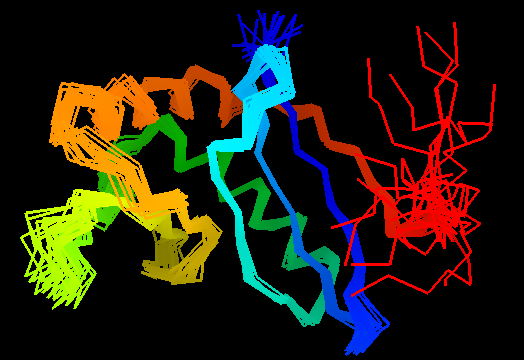
T0472 (2k49): UPF0339 protein SO3888 from Shewanella oneidensis
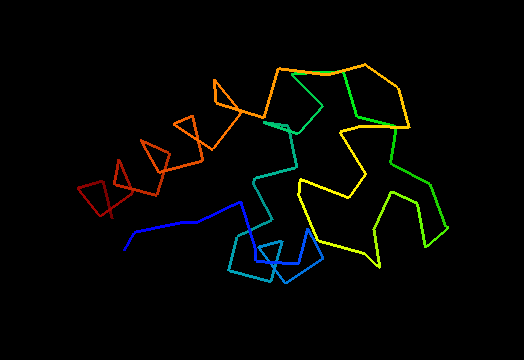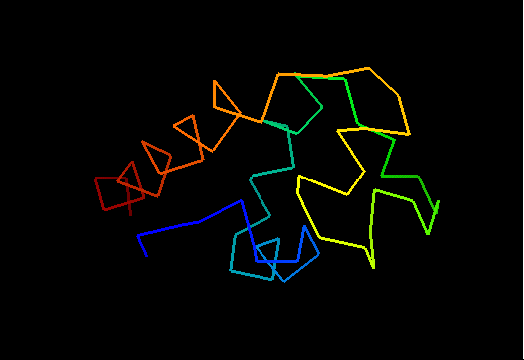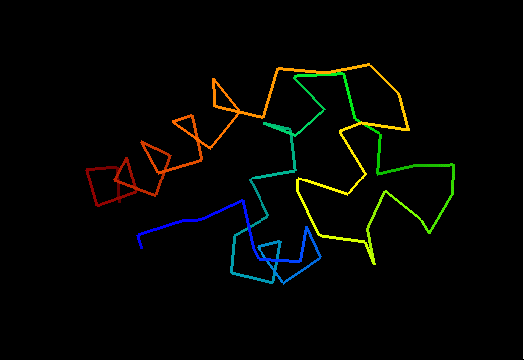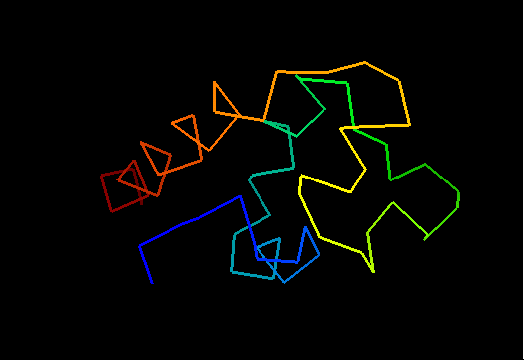
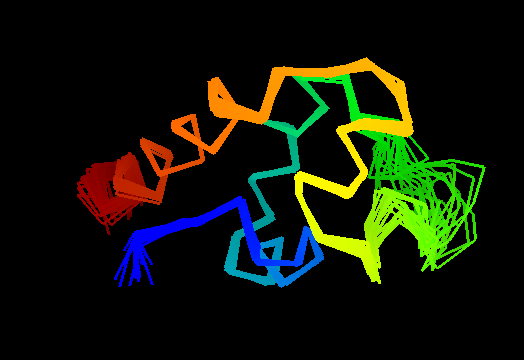
T0473 (2k53): A3DK08 protein from Clostridium thermocellum
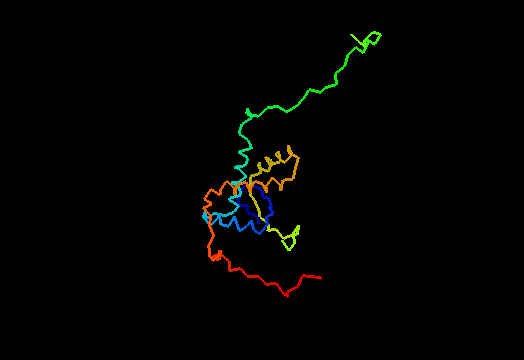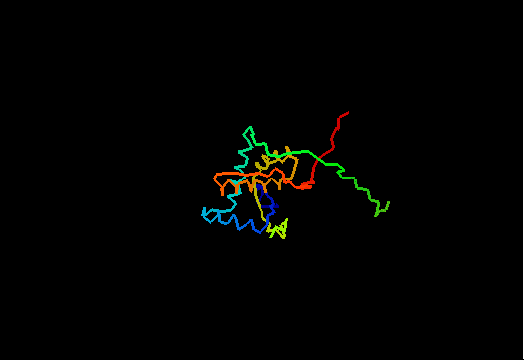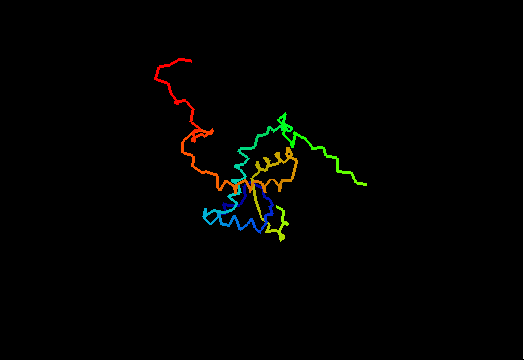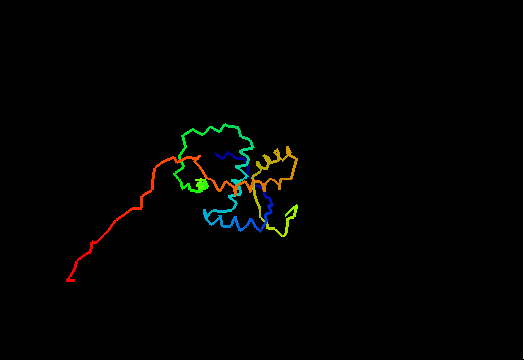
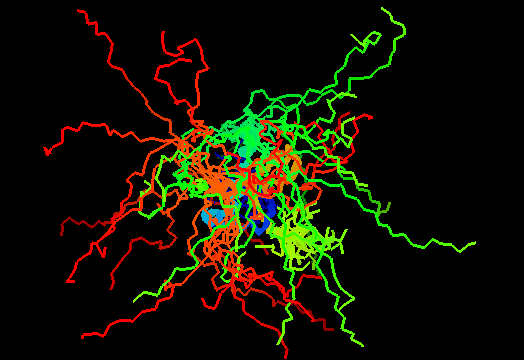
T0474 (2k5j): yiiF from Shigella flexneri serotype 5b (strain 8401)
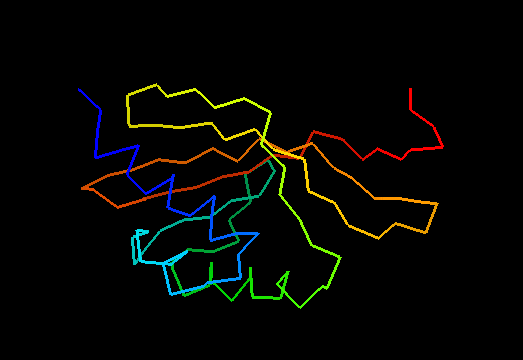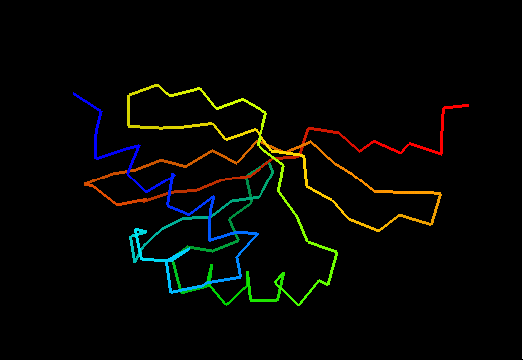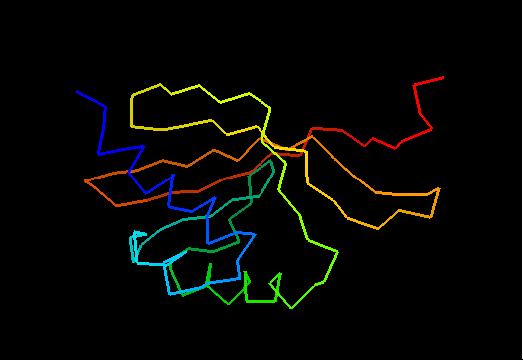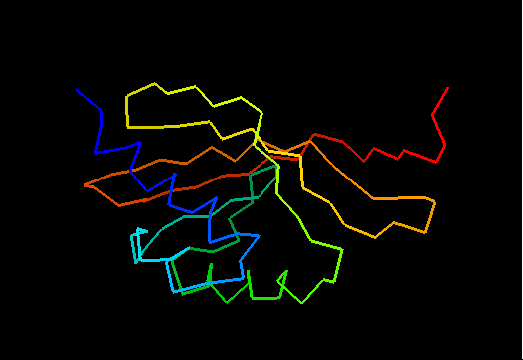
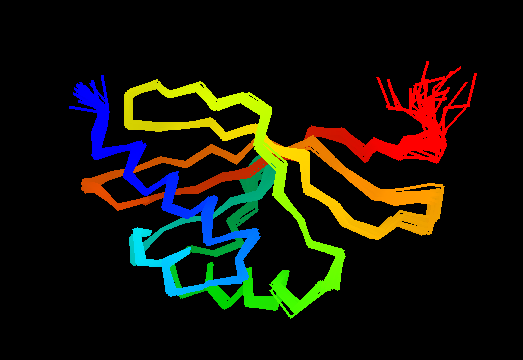
T0475 (2k54): protein Atu0742 from Agrobacterium tumefaciens. This structure is quite good, actually!
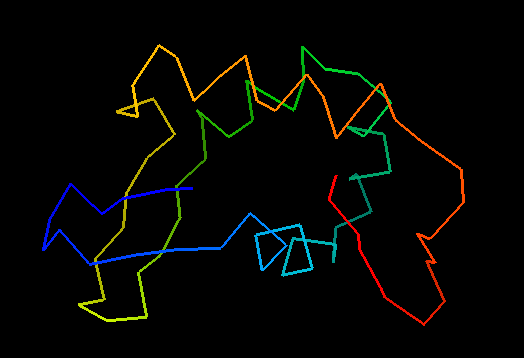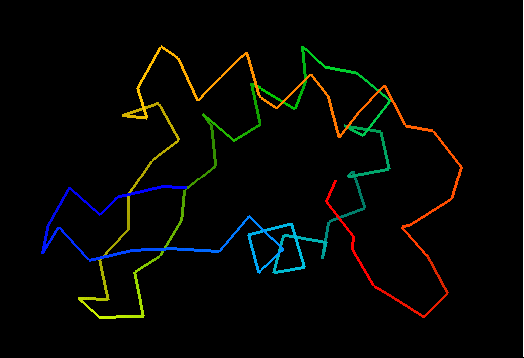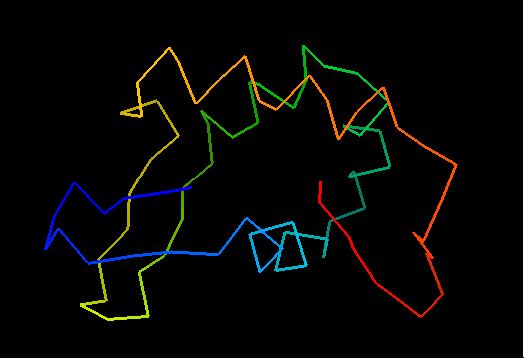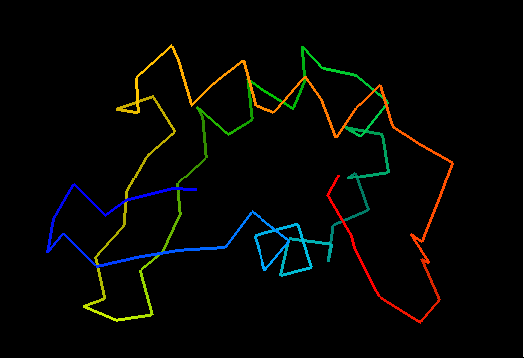
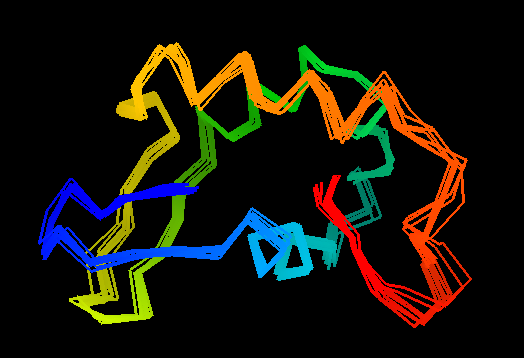
T0476 (2k5c): protein PF0385 from Pyrococcus furiosus. Yet another good structure. NMR folks can do something right once in a while!
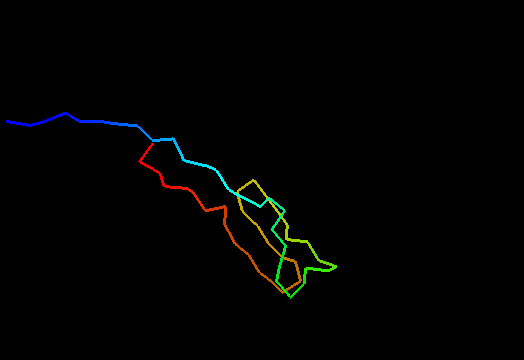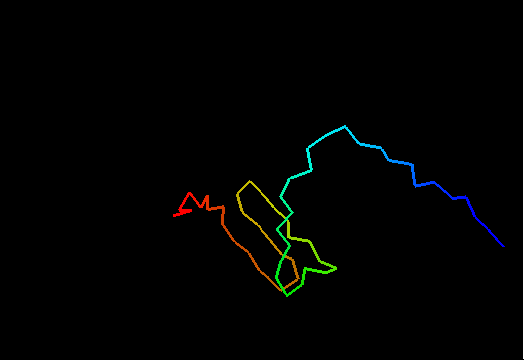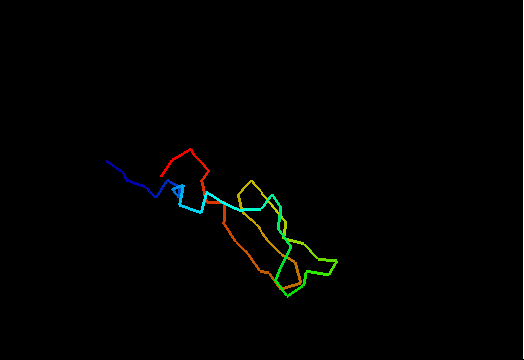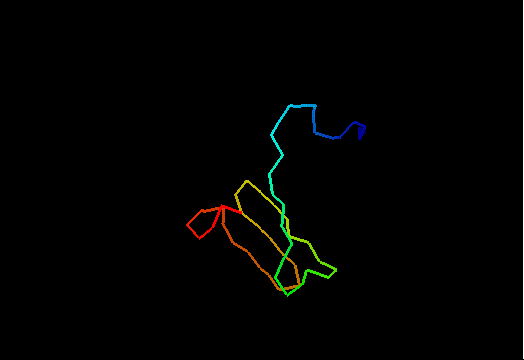
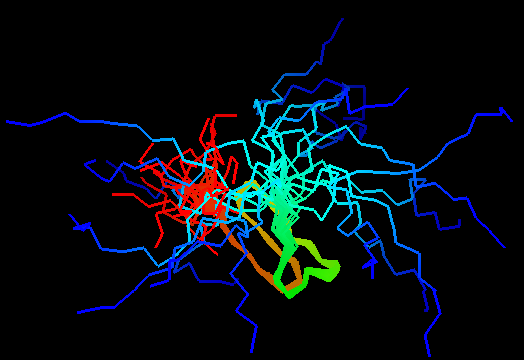
T0480 (2k4x): 30S ribosomal protein S27A from Thermoplasma acidophilum
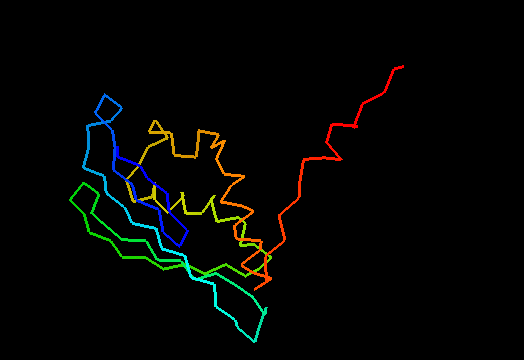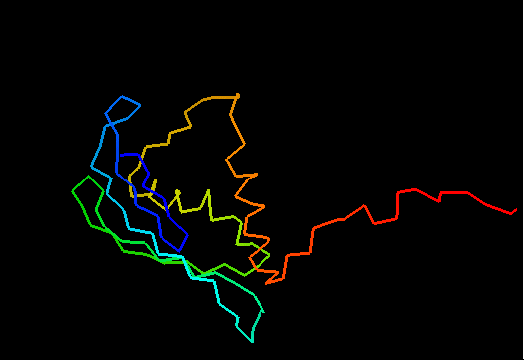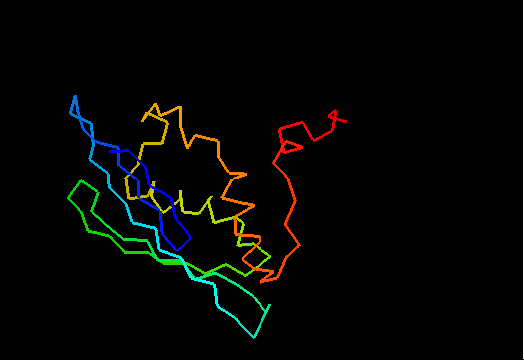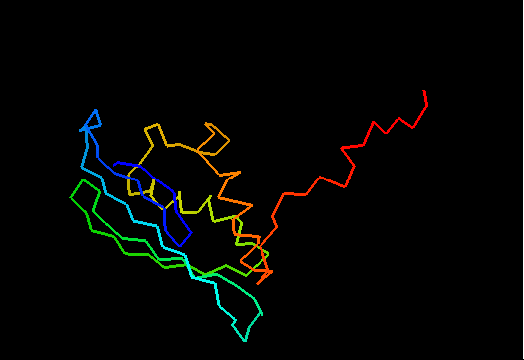
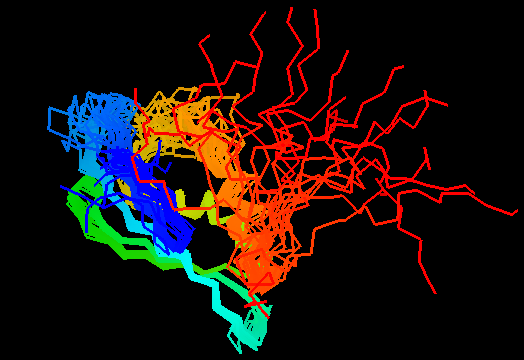
T0482 (2k4v): protein PA1076 from Pseudomonas aeruginosa

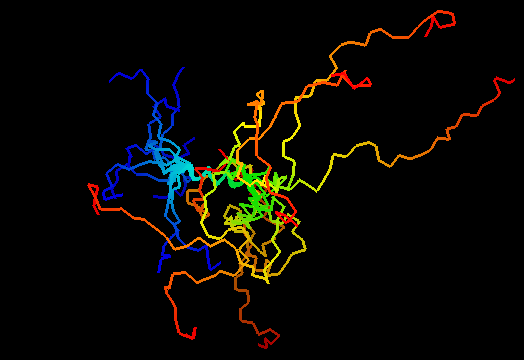
T0484 (2k5k): RhR2 from Rhodobacter sphaeroides
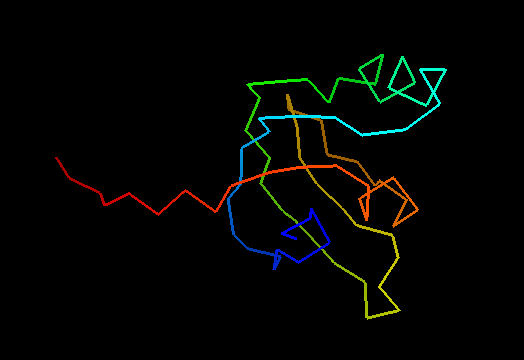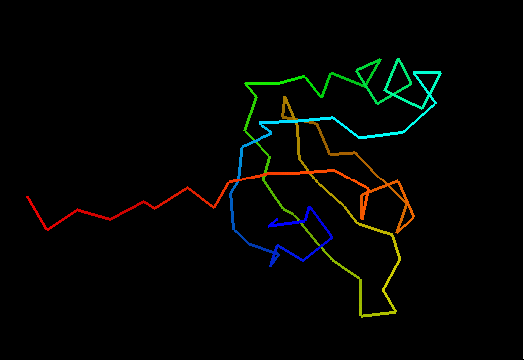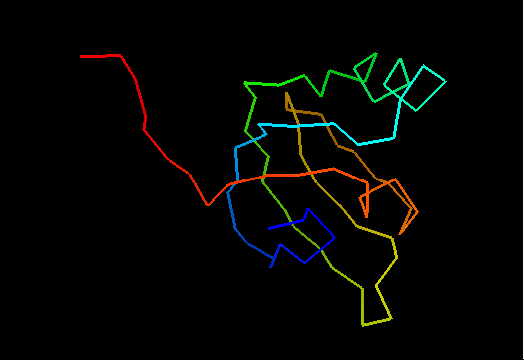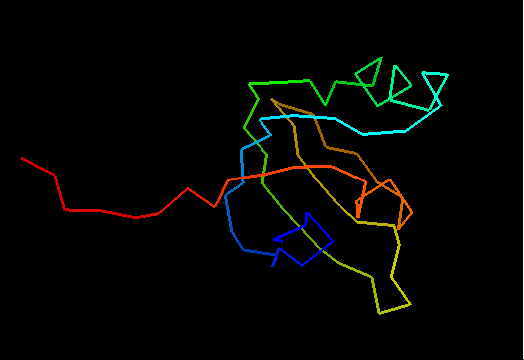
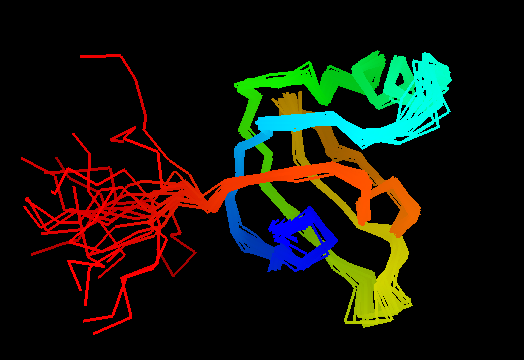
T0492 (2k5l): protein FeoA from Clostridium thermocellum
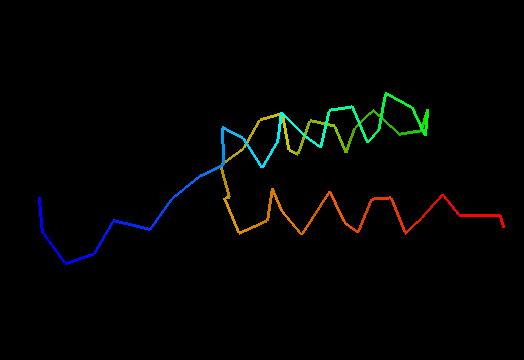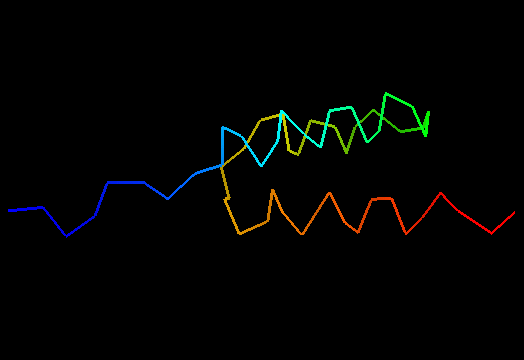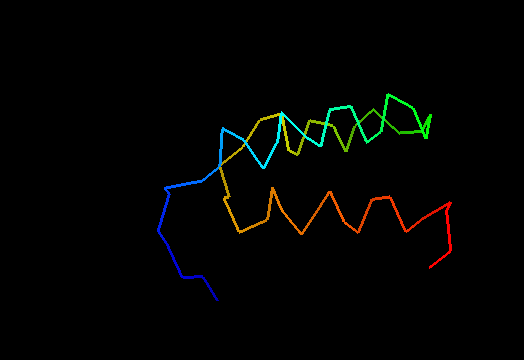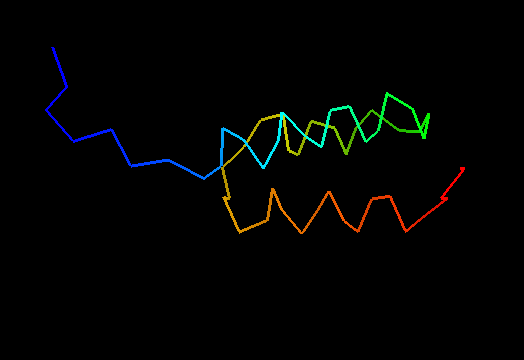
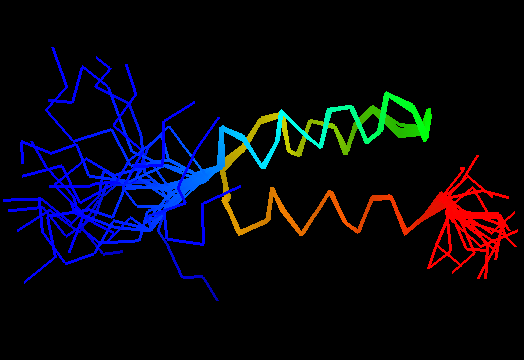
T0498 (2jws, not target but close enough): in-vitro selected protein L1c
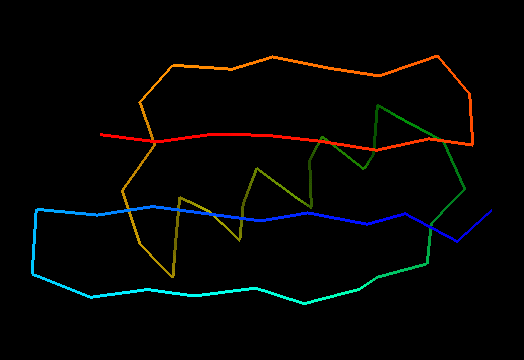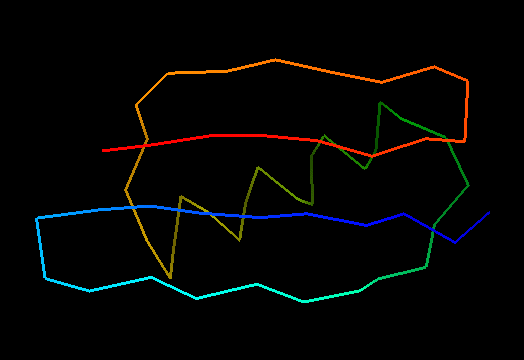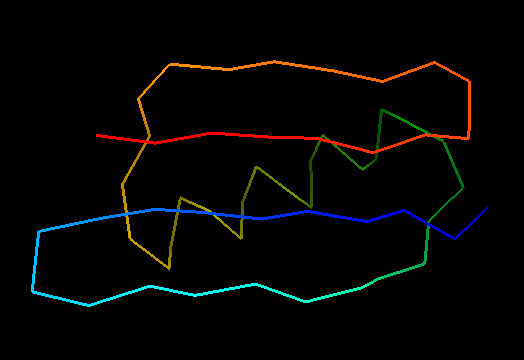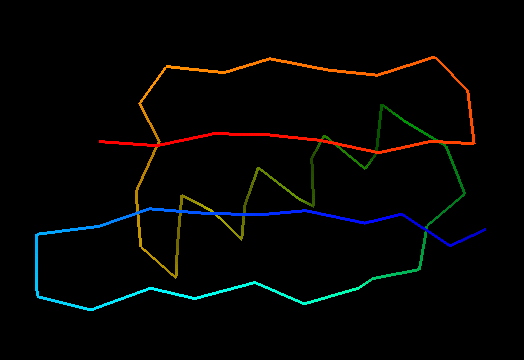
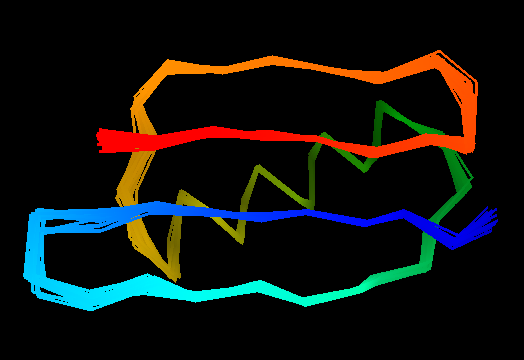
T0499 (2jwu, not target but close enough): in-vitro selected protein L1d
Claimed to be disordered