T0466
protein SAG0934 from Streptococcus agalactiae
Target sequence:
>T0466 SaR32, Streptococcus agalactiae, 128 residues
MMRLANGIVLDKDTTFGELKFSALRREVRIQNEDGSVSDEIKERTYDLKSKGQGRMIQVSIPASVPLKEFDYNARVELINPIADTVATATYQGADVDWYIKADDIVLTKDSSSFKAQPQAKKEPTQDK
Structure:
Method: NMR
Determined by: NESG
PDB ID: 2k5d
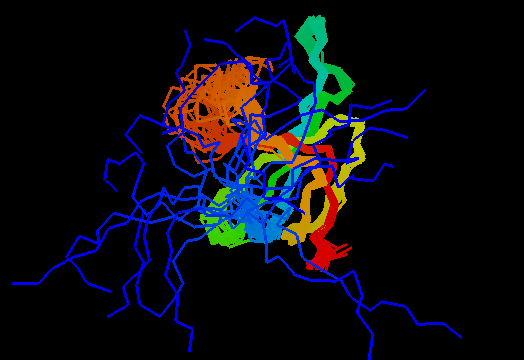
Domains: PyMOL of domains
Single domain protein with poorly structured N-terminus and a loop. Structurally variable regions were removed from NMR models for evaluation of predictions.
Structure classification:
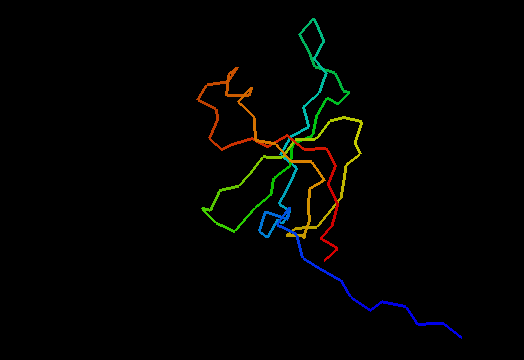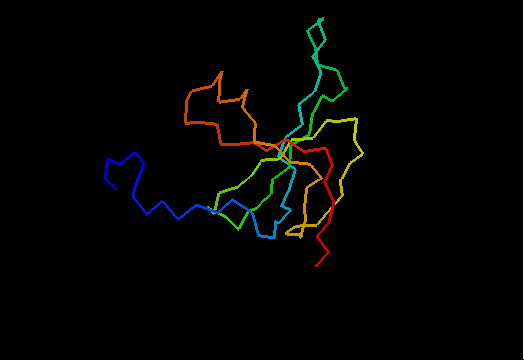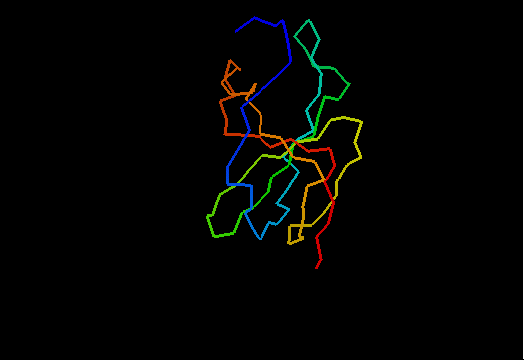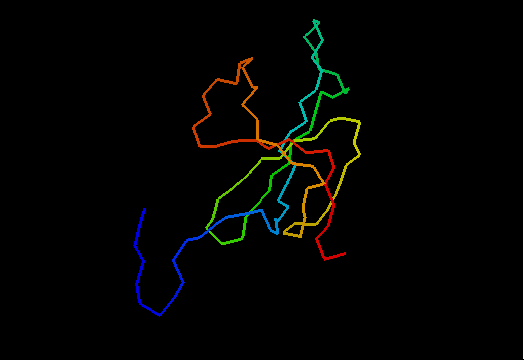
OB-fold. 5-stranded β-barrel. The five strands are shown in the ribbon diagram below as dark blue, cyan-green, lime-green, yellow and red. In addition to the 5-stranded OB-core, this structure is elaborated with a long insertion between β-strands 4 and 5 housing an α-helix. The two β-strands (orange-yellow and orange) in this region are extensions of the β-strands 4 and 5 and they close the barrel.
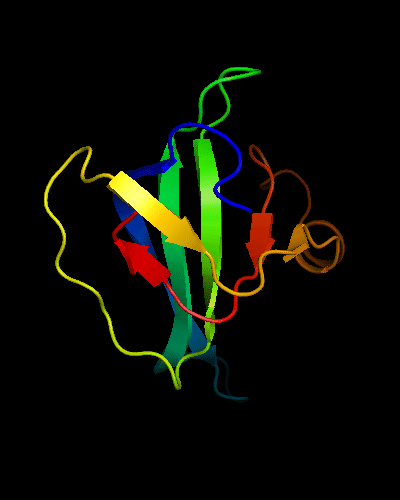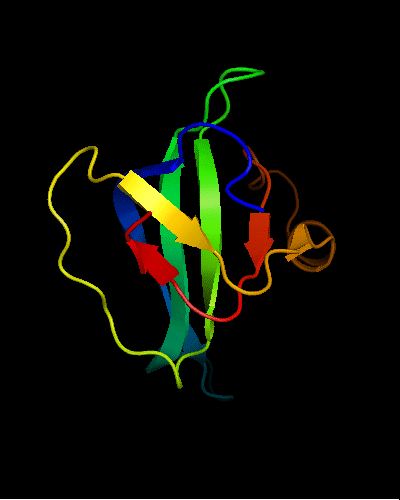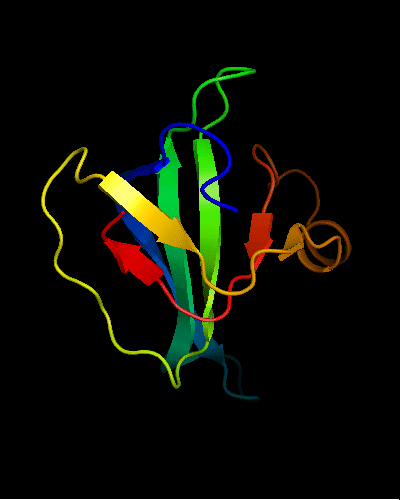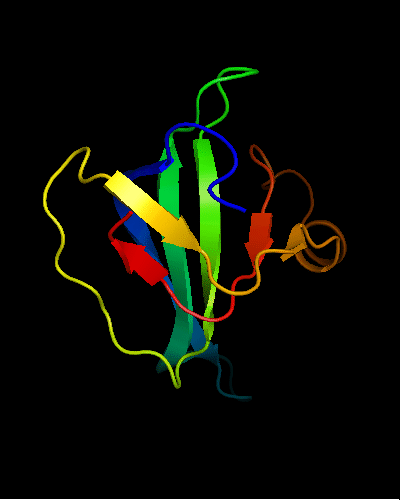
Cartoon diagram of T0466: 2k5d model 1 residues 14-108
CASP category:
Fold recognition. Fold similarity not detectable statistically by sequence methods known to us.
Closest templates:
All three COMPASS hit to OB-fold proteins are to the Nucleic acid - binding domain superfamily, however, alignments are wrong as they do not validate structurally. First hit is #27 in the total list of hits and its E-value is around 35, so it can hardy be called statistically significant in any case. Moreover, proteins from all SCOP classes were found by COMPASS prior to this hit. According to structure, proteins from 'Nucleic acid - binding' superfamily are also most similar to T0466.
Target sequence - PDB file inconsistencies:
NMR models contain two C-terminal residues 109-LE-110 not present in the target sequence. Instead of these two residues, target sequence contains the following C-terminus 109-KDSSSFKAQPQAKKEPTQDK-128 apparently cloned out for structure determination.
T0466 2k5d.pdb T0466.pdb PyMOL PyMOL of domains
T0466 1 MMRLANGIVLDKDTTFGELKFSALRREVRIQNEDGSVSDEIKERTYDLKSKGQGRMIQVSIPASVPLKEFDYNARVELINPIADTVATATYQGADVDWYIKADDIVLTKDSSSFKAQPQAKKEPTQDK-- 128 *************||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||********|||||||||||||~~~~~~~~~~~~~~~~~~~~~~ 2k5dA 1 MMRLANGIVLDKDTTFGELKFSALRREVRIQNEDGSVSDEIKERTYDLKSKGQGRMIQVSIPASVPLKEFDYNARVELINPIADTVATATYQGADVDWYIKADDIVLT--------------------LE 110
Residue change log: remove 109-LE-110 as it is not present in target sequence; remove 1-MMRLANGIVLDKD-13, 88-TATYQGAD-95 by 3.5Å SD cutoff;
Single domain protein: target 14-87, 96-108 ; pdb 14-87, 96-108
Sequence classification:
DUF961 in Pfam, which includes just a few very close homologs. Below is PROMALS3D alignment of several most distant homologs detectable by PSI-BLAST.
Conservation 6 5 978 5 6 7 657 5 7 8 58 5 5 6 66 8 655 188584883 TYIEQVERVVARVDITGTVDYLEKGIVVDIYDENG-----------EIMEELEVS----------PGMVEVKIPMNYPEK----------EVPV-------EVDIEDNLP---EFYLESVEISPEQVEVGGWESVIN--------------
29374784 MGLTFENNTIENFDVQATFGMMNFLEVVAINERDGE---------NNVTEEVREQRVTVYSSKL-NDQVEIVIDPDYDVS----GINYDDEIELTGNVTARGWLYTFKGYNDNVQTEQAFKVRAAGIKKVGAVKQTQAPQPKEKQENKQ--
29377029 MGLTFENNTIENFDVQGTFGTMNFLEVVAINERDGE---------GNVTDEVREQRVTVYSSKL-NDQVEIAIDPDYDVS----GINYDDEIELTGNVTARGWLNTFKGFNDNVQTEQAFKVRSAGIKKVGAVKPTQTPQPREKQENK---116872189 MSLKFEENTIKDFDVKKTFGTCYYLSAQSIYENDSE---------GNRTEIVREQRVTIYSEVK-NDQIDITLPETTDLA----SFSYDDEIELTGEVTALAWLSSYAGYN--DSIQSEQAFKIRAGGIKKVTTPHQKPEKQKSTGTLQGE
49483485 --------MKPVLDIKKTLGQLNFLGVDEKYKYEN-----------GERTDKTVYAYKLASQEQ-GEQITVKTPNKVE-------LDYLQECELVEPDVKMYVQMSGDF------GTIAYSWNAEDIKVVGSNSALKQK------------
24378720 MTLKYAADVIEKFDLDKTFGTLNFLETVPIMKWEDYLDESTGEEKRRETDELEALDVKIYSSAA-GGIITVTVPPEAKVIQLTPEKNYNDEVKLVAPTARFWSNSEVINGR--RVVTSGVKIRAKDVVVSNVGKSNTKEIKPDK-------
167767490 ----MRLSNGFVIDKEKTFGELKFTAVR-DVFLQNED--------GTPSTQLKKRIYDLKCSLH-GGIIPVSVPPEVPLR----EFPYNAVVELVNPVADTVSRKTYTG------ADVDWYVKAEDIVLKNKGNQNAGSPQNHTPQGQPKK
126700965 ----MRLANGIVIDKEATFGALKFSALRREVHLQNED--------GSVSEEIKERTYDLKSRGQ-GRMIQVSIPASVPLK----EFDYNAEVELINPVADTVATATFQG------AEVDWYIKAEDIVLKKGTSMNPQPPKKDTPPVK---
194271962 ----MRLAEGMIINQEKTFGTLKFSALRREVRKQNED--------GTVSTEIKERTYNLKSSTQ-GRMIQVSIPAEVPLR----EFPYDAEVELVQPTVDTVANYVFREG-----TTVSWFIKAEDLILKKNGPVSQQQEKPVPKK-----153174536 ----MKFTEKVKLDIAATFGTLEFLGHDGERTKAGE----------DNQSVVLNQRYGLSSSAQ-EGTVVIYLPADKKLS-----FDYDTEVTLVNPTISAFATATYNA------MDVITQIFADDIIPVAKNQTNTSHKGKEAN------
153174537 -----MELKF-IATTNETFENVRYAGHDATKDIKTG----------FGRNEKLEKRYFALLSQDFPGGLEVYLPEVAGEK----DFAFKTKVKLVNPVMIPVASRIGNNN-----ALVNWELHADDLVPFK--------------------
194271961 -----MELKFVVPNMEKTFGHLTYAGEGEVLTEGS-----------GRNRVVIGRSYHLYSDIQRADDIEIIVAPEVGEK----IFKDGELVKVVNPKIQVEGYQIDNR------GYTNYLLKVDDLVKVNGGM-----------------
126700966 -----MELKFVIPNMEKTFGNLEFAGEDKTEQRRI-----------NGRMAVLSRSFNLYSDVQRADDIVVILPAEAGEK----HFDFEERVKLVNPRITAEGYKIGTR------GFTNYILHADDMVKA---------------------
167767710 -----MEMKYVVPDMAQSFGTLEFAGESEPVFERDK----------NNRRVLTRRSYNLYSDVQKGENVVVEIPVQAGEK----HFKYEQKVKLVNPKLYGRGYAIGDM------GHTDYVLVADDIVAVEEK------------------
16077552 -----MNLNFIVPDINMTFGDMKFMGLNRERYVYDRE-------NNKRTDVLESRIYNIASAVQ-GGQIEVTIPEYAGAK----EIPPFADIELKNPKISAMATSQRDST----YANVMWKLEADDIVVKGGSSVKPAAATGGNEKK----
166033406 -----MQITLTKEQVKEAFGELRFMGADNCYRYDKDS--------KKKTEEVEALKLHLGSMKL-GNSVDIRLEAT-ELP----KIEPYAVVELEEPVYAPYVQRGNF-------PVLVEKITCKGIRPVANKNM----------------
T0466 ---MMRLANGIVLDKDTTFGELKFSALRREVRIQNED--------GSVSDEIKERTYDLKSKGQ-GRMIQVSIPASVPLK----EFDYNARVELINPIADTVATATYQG------ADVDWYIKADDIVLTKDSSSFKAQPQAKKEPTQDK-Consensus_aa ch...........s.p.TFGplp@.............p........p.pp.l..pp@plbtp.p.s..l.V.lP.phsbpp....h.hp.clcllsP.h.h.s.......p....s..sh.l.h-sh........................
Consensus_ss ee ee hhh ee ee eee eeeeeeeeeee eeeeee eeee eeeeeeee e eeeeeeeee eeeee
Comments:
ROSETTA failed to predict this OB-fold de-novo. Metaserver bioinfo.pl did not find similarity to other OB-fold proteins. Although some servers used OB-folds as templates, it is unclear how significant those predictions are, as quite a few all-β folds were used as templates, in particular, lipocalins, also a β-barrel fold. The bias towards lipocalin fold in ab-initio servers is likely caused by the local meander-like nature β-strand interactions in it.
Server predictions:
T0466:pdb 14-108:seq 14-108:FR; alignment
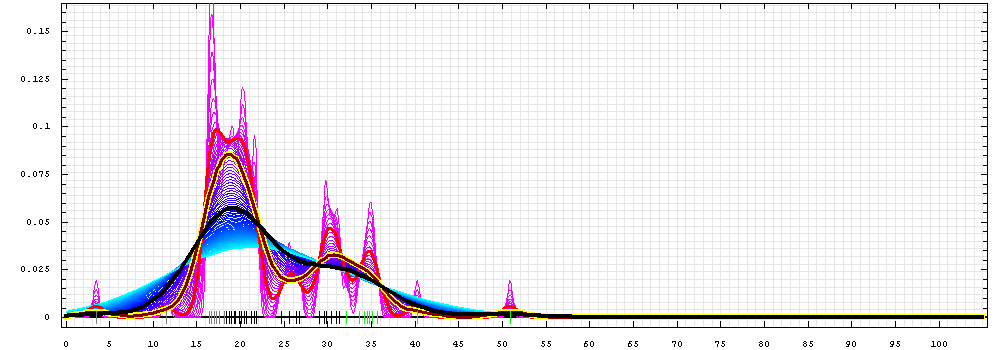
First models for T0466:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0466 | T0466 | T0466 | T0466 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | MUSTER | 50.86 | 44.16 | 49.12 | 3.98 | 4.26 | 2.89 | 50.86 | 44.16 | 49.12 | 2.64 | 2.74 | 2.23 | MUSTER | 1 |
| 2 | ZicoFullSTP | 48.56 | 42.15 | 46.87 | 3.62 | 3.89 | 2.58 | 48.56 | 42.91 | 46.87 | 2.36 | 2.56 | 1.93 | ZicoFullSTP | 2 |
| 3 | Chicken_George | 44.25 | 39.08 | 44.31 | 2.95 | 3.32 | 2.22 | 45.98 | 40.04 | 45.63 | 2.04 | 2.15 | 1.77 | Chicken_George | 3 |
| 4 | BAKER−ROBETTA | 40.23 | 32.95 | 39.65 | 2.33 | 2.19 | 1.58 | 40.23 | 32.95 | 39.65 | 1.34 | 1.14 | 0.99 | BAKER−ROBETTA | 4 |
| 5 | 3DShotMQ | 37.93 | 25.86 | 39.79 | 1.97 | 0.88 | 1.60 | 37.93 | 25.86 | 39.79 | 1.05 | 0.13 | 1.01 | 3DShotMQ | 5 |
| 6 | MULTICOM−REFINE | 35.63 | 28.35 | 39.68 | 1.61 | 1.34 | 1.58 | 35.63 | 29.02 | 39.95 | 0.77 | 0.58 | 1.03 | MULTICOM−REFINE | 6 |
| 7 | Elofsson | 35.63 | 28.35 | 39.59 | 1.61 | 1.34 | 1.57 | 35.63 | 28.35 | 39.59 | 0.77 | 0.49 | 0.99 | Elofsson | 7 |
| 8 | MUProt | 35.06 | 27.97 | 39.64 | 1.52 | 1.27 | 1.58 | 35.06 | 29.60 | 39.64 | 0.70 | 0.66 | 0.99 | MUProt | 8 |
| 9 | Bates_BMM | 35.06 | 27.59 | 39.94 | 1.52 | 1.20 | 1.62 | 35.06 | 29.41 | 39.99 | 0.70 | 0.64 | 1.04 | Bates_BMM | 9 |
| 10 | MULTICOM−CLUSTER | 35.06 | 27.20 | 39.51 | 1.52 | 1.13 | 1.56 | 35.06 | 29.41 | 39.51 | 0.70 | 0.64 | 0.98 | MULTICOM−CLUSTER | 10 |
| 11 | GS−KudlatyPred | 34.77 | 25.57 | 39.19 | 1.48 | 0.83 | 1.51 | 38.51 | 32.95 | 40.72 | 1.12 | 1.14 | 1.13 | GS−KudlatyPred | 11 |
| 12 | MULTICOM−RANK | 34.48 | 31.70 | 40.18 | 1.43 | 1.96 | 1.65 | 34.48 | 31.70 | 40.18 | 0.63 | 0.96 | 1.06 | MULTICOM−RANK | 12 |
| 13 | MULTICOM−CMFR | 34.20 | 29.69 | 39.93 | 1.39 | 1.59 | 1.62 | 34.20 | 29.69 | 39.93 | 0.60 | 0.68 | 1.03 | MULTICOM−CMFR | 13 |
| 14 | Zhang | 33.91 | 28.45 | 37.20 | 1.34 | 1.36 | 1.24 | 37.93 | 28.45 | 37.34 | 1.05 | 0.50 | 0.69 | Zhang | 14 |
| 15 | Zhang−Server | 33.62 | 26.34 | 35.55 | 1.30 | 0.97 | 1.01 | 33.62 | 26.34 | 35.55 | 0.52 | 0.20 | 0.46 | Zhang−Server | 15 |
| 16 | ABIpro | 32.18 | 27.97 | 35.48 | 1.07 | 1.27 | 1.00 | 32.18 | 27.97 | 35.48 | 0.35 | 0.43 | 0.45 | ABIpro | 16 |
| 17 | FALCON | 32.18 | 27.97 | 35.48 | 1.07 | 1.27 | 1.00 | 37.64 | 32.09 | 39.27 | 1.02 | 1.02 | 0.94 | FALCON | 17 |
| 18 | KudlatyPredHuman | 31.90 | 29.21 | 38.71 | 1.03 | 1.50 | 1.45 | 35.34 | 29.31 | 39.74 | 0.74 | 0.62 | 1.01 | KudlatyPredHuman | 18 |
| 19 | MULTICOM | 31.90 | 29.21 | 40.58 | 1.03 | 1.50 | 1.71 | 38.79 | 33.81 | 41.43 | 1.16 | 1.26 | 1.23 | MULTICOM | 19 |
| 20 | GeneSilico | 31.90 | 26.82 | 31.46 | 1.03 | 1.06 | 0.44 | 31.90 | 26.82 | 32.11 | 0.31 | 0.27 | 0.01 | GeneSilico | 20 |
| 21 | McGuffin | 31.61 | 29.31 | 39.74 | 0.99 | 1.52 | 1.59 | 37.64 | 31.70 | 39.74 | 1.02 | 0.96 | 1.01 | McGuffin | 21 |
| 22 | mufold | 31.61 | 28.54 | 38.86 | 0.99 | 1.38 | 1.47 | 34.48 | 28.54 | 38.86 | 0.63 | 0.51 | 0.89 | mufold | 22 |
| 23 | Jones−UCL | 31.61 | 23.95 | 34.79 | 0.99 | 0.53 | 0.90 | 35.34 | 32.47 | 41.03 | 0.74 | 1.07 | 1.17 | Jones−UCL | 23 |
| 24 | ZicoFullSTPFullData | 31.32 | 29.69 | 38.59 | 0.94 | 1.59 | 1.43 | 48.56 | 42.15 | 46.87 | 2.36 | 2.45 | 1.93 | ZicoFullSTPFullData | 24 |
| 25 | FALCON_CONSENSUS | 31.32 | 27.49 | 33.77 | 0.94 | 1.18 | 0.76 | 37.64 | 31.70 | 39.27 | 1.02 | 0.96 | 0.94 | FALCON_CONSENSUS | 25 |
| 26 | PSI | 31.03 | 27.78 | 38.29 | 0.90 | 1.24 | 1.39 | 31.61 | 29.31 | 39.74 | 0.28 | 0.62 | 1.01 | PSI | 26 |
| 27 | MidwayFolding | 31.03 | 23.56 | 34.51 | 0.90 | 0.46 | 0.86 | 31.03 | 23.56 | 34.51 | 0.21 | 0.32 | MidwayFolding | 27 | |
| 28 | RAPTOR | 31.03 | 23.56 | 34.51 | 0.90 | 0.46 | 0.86 | 31.03 | 24.14 | 34.51 | 0.21 | 0.32 | RAPTOR | 28 | |
| 29 | PS2−server | 30.46 | 25.10 | 31.78 | 0.81 | 0.74 | 0.49 | 30.46 | 25.10 | 31.78 | 0.14 | 0.02 | PS2−server | 29 | |
| 30 | PS2−manual | 30.46 | 25.10 | 31.78 | 0.81 | 0.74 | 0.49 | 30.46 | 25.10 | 31.78 | 0.14 | 0.02 | PS2−manual | 30 | |
| 31 | fais−server | 30.46 | 24.23 | 32.76 | 0.81 | 0.58 | 0.62 | 30.46 | 24.23 | 32.76 | 0.14 | 0.10 | fais−server | 31 | |
| 32 | fams−ace2 | 30.17 | 26.92 | 37.98 | 0.76 | 1.08 | 1.35 | 34.77 | 29.41 | 39.93 | 0.67 | 0.64 | 1.03 | fams−ace2 | 32 |
| 33 | GS−MetaServer2 | 29.89 | 23.37 | 28.69 | 0.72 | 0.42 | 0.06 | 29.89 | 23.37 | 28.69 | 0.07 | GS−MetaServer2 | 33 | ||
| 34 | GeneSilicoMetaServer | 29.89 | 23.37 | 28.69 | 0.72 | 0.42 | 0.06 | 29.89 | 23.37 | 28.69 | 0.07 | GeneSilicoMetaServer | 34 | ||
| 35 | FrankensteinLong | 29.60 | 25.00 | 31.43 | 0.67 | 0.72 | 0.44 | 29.60 | 25.00 | 31.43 | 0.03 | 0.01 | FrankensteinLong | 35 | |
| 36 | Softberry | 29.60 | 24.62 | 30.22 | 0.67 | 0.65 | 0.27 | 29.60 | 24.62 | 30.22 | 0.03 | Softberry | 36 | ||
| 37 | MUFOLD−MD | 29.60 | 24.43 | 36.14 | 0.67 | 0.62 | 1.09 | 29.60 | 24.43 | 36.14 | 0.03 | 0.54 | MUFOLD−MD | 37 | |
| 38 | METATASSER | 29.60 | 24.23 | 33.79 | 0.67 | 0.58 | 0.76 | 40.80 | 36.40 | 36.95 | 1.41 | 1.63 | 0.64 | METATASSER | 38 |
| 39 | FAMS−multi | 29.31 | 22.32 | 31.96 | 0.63 | 0.23 | 0.51 | 30.17 | 25.57 | 33.51 | 0.10 | 0.09 | 0.19 | FAMS−multi | 39 |
| 40 | 3DShot1 | 29.31 | 22.03 | 34.36 | 0.63 | 0.17 | 0.84 | 29.31 | 22.03 | 34.36 | 0.31 | 3DShot1 | 40 | ||
| 41 | Kolinski | 29.02 | 25.00 | 30.85 | 0.58 | 0.72 | 0.36 | 30.17 | 25.00 | 30.85 | 0.10 | 0.01 | Kolinski | 41 | |
| 42 | Phyre2 | 29.02 | 23.85 | 29.74 | 0.58 | 0.51 | 0.20 | 29.02 | 23.85 | 31.64 | Phyre2 | 42 | |||
| 43 | Sasaki−Cetin−Sasai | 28.16 | 22.99 | 30.35 | 0.45 | 0.35 | 0.29 | 28.16 | 22.99 | 30.51 | Sasaki−Cetin−Sasai | 43 | |||
| 44 | TASSER | 26.72 | 21.74 | 27.74 | 0.23 | 0.12 | 26.72 | 22.03 | 43.09 | 1.44 | TASSER | 44 | |||
| 45 | pro−sp3−TASSER | 26.72 | 21.74 | 27.74 | 0.23 | 0.12 | 46.55 | 40.61 | 45.98 | 2.11 | 2.23 | 1.82 | pro−sp3−TASSER | 45 | |
| 46 | JIVE08 | 26.44 | 25.29 | 27.16 | 0.18 | 0.78 | 26.44 | 25.29 | 27.16 | 0.05 | JIVE08 | 46 | |||
| 47 | BioSerf | 26.44 | 20.50 | 32.24 | 0.18 | 0.55 | 27.87 | 24.81 | 37.81 | 0.75 | BioSerf | 47 | |||
| 48 | POEM | 26.44 | 19.92 | 29.58 | 0.18 | 0.18 | 28.74 | 24.33 | 34.26 | 0.29 | POEM | 48 | |||
| 49 | PRI−Yang−KiharA | 25.86 | 22.41 | 30.18 | 0.09 | 0.24 | 0.27 | 25.86 | 22.41 | 30.18 | PRI−Yang−KiharA | 49 | |||
| 50 | DBAKER | 25.57 | 21.93 | 30.11 | 0.05 | 0.15 | 0.26 | 25.57 | 24.14 | 36.73 | 0.61 | DBAKER | 50 | ||
| 51 | Poing | 25.57 | 20.21 | 32.45 | 0.05 | 0.58 | 25.57 | 20.21 | 32.45 | 0.06 | Poing | 51 | |||
| 52 | Ozkan−Shell | 24.71 | 20.31 | 30.27 | 0.28 | 37.07 | 32.28 | 39.23 | 0.95 | 1.04 | 0.94 | Ozkan−Shell | 52 | ||
| 53 | SHORTLE | 24.71 | 20.11 | 24.65 | 27.87 | 24.62 | 25.66 | SHORTLE | 53 | ||||||
| 54 | SAM−T08−server | 24.71 | 19.73 | 28.53 | 0.04 | 24.71 | 20.21 | 28.53 | SAM−T08−server | 54 | |||||
| 55 | SAM−T08−human | 24.43 | 19.44 | 26.55 | 39.37 | 33.24 | 41.75 | 1.23 | 1.18 | 1.27 | SAM−T08−human | 55 | |||
| 56 | AMU−Biology | 23.85 | 21.36 | 27.37 | 0.05 | 35.34 | 32.85 | 34.25 | 0.74 | 1.13 | 0.29 | AMU−Biology | 56 | ||
| 57 | fais@hgc | 23.85 | 21.26 | 28.31 | 0.03 | 0.01 | 31.03 | 21.26 | 33.32 | 0.21 | 0.17 | fais@hgc | 57 | ||
| 58 | LEE | 23.85 | 19.64 | 29.93 | 0.23 | 27.59 | 22.80 | 31.80 | LEE | 58 | |||||
| 59 | Hao_Kihara | 23.56 | 18.77 | 24.82 | 38.51 | 32.57 | 35.61 | 1.12 | 1.09 | 0.47 | Hao_Kihara | 59 | |||
| 60 | Scheraga | 23.28 | 18.49 | 23.92 | 23.85 | 20.40 | 28.78 | Scheraga | 60 | ||||||
| 61 | SMEG−CCP | 23.28 | 17.62 | 35.02 | 0.94 | 23.28 | 17.62 | 35.02 | 0.39 | SMEG−CCP | 61 | ||||
| 62 | POISE | 22.99 | 18.97 | 21.38 | 23.28 | 19.06 | 26.17 | POISE | 62 | ||||||
| 63 | Wolynes | 22.70 | 21.17 | 23.19 | 0.01 | 23.85 | 21.17 | 27.98 | Wolynes | 63 | |||||
| 64 | taylor | 22.70 | 18.97 | 25.94 | 22.70 | 18.97 | 25.94 | taylor | 64 | ||||||
| 65 | Sternberg | 22.41 | 20.11 | 24.86 | 22.41 | 20.11 | 27.86 | Sternberg | 65 | ||||||
| 66 | ProteinShop | 22.13 | 17.72 | 27.60 | 30.46 | 26.63 | 30.57 | 0.14 | 0.24 | ProteinShop | 66 | ||||
| 67 | Phyre_de_novo | 21.84 | 19.73 | 30.10 | 0.25 | 21.84 | 19.73 | 30.10 | Phyre_de_novo | 67 | |||||
| 68 | FAMSD | 21.84 | 18.01 | 23.27 | 26.44 | 22.03 | 29.31 | FAMSD | 68 | ||||||
| 69 | mariner1 | 21.55 | 19.54 | 22.49 | 50.86 | 44.73 | 46.74 | 2.64 | 2.82 | 1.92 | mariner1 | 69 | |||
| 70 | MeilerLabRene | 21.55 | 18.30 | 21.81 | 21.84 | 18.49 | 24.87 | MeilerLabRene | 70 | ||||||
| 71 | ACOMPMOD | 21.55 | 18.10 | 20.89 | 22.99 | 18.10 | 20.89 | ACOMPMOD | 71 | ||||||
| 72 | circle | 21.55 | 16.38 | 23.83 | 26.44 | 22.03 | 29.31 | circle | 72 | ||||||
| 73 | Zico | 21.26 | 19.64 | 26.80 | 48.56 | 42.15 | 46.87 | 2.36 | 2.45 | 1.93 | Zico | 73 | |||
| 74 | HHpred2 | 21.26 | 16.86 | 21.39 | 21.26 | 16.86 | 21.39 | HHpred2 | 74 | ||||||
| 75 | TJ_Jiang | 20.98 | 19.83 | 22.50 | 21.84 | 20.79 | 28.29 | TJ_Jiang | 75 | ||||||
| 76 | Bilab−UT | 20.98 | 17.72 | 32.25 | 0.55 | 27.01 | 22.80 | 33.35 | 0.17 | Bilab−UT | 76 | ||||
| 77 | fleil | 20.98 | 17.53 | 21.22 | 22.41 | 18.97 | 21.44 | fleil | 77 | ||||||
| 78 | Handl−Lovell | 20.69 | 18.39 | 25.21 | 23.56 | 20.50 | 28.75 | Handl−Lovell | 78 | ||||||
| 79 | FEIG | 20.69 | 17.05 | 23.41 | 24.43 | 21.74 | 24.12 | FEIG | 79 | ||||||
| 80 | FLOUDAS | 20.69 | 16.95 | 23.80 | 24.43 | 22.22 | 30.07 | FLOUDAS | 80 | ||||||
| 81 | CpHModels | 20.69 | 16.86 | 21.46 | 20.69 | 16.86 | 21.46 | CpHModels | 81 | ||||||
| 82 | Distill | 20.69 | 14.94 | 25.91 | 22.41 | 18.20 | 28.53 | Distill | 82 | ||||||
| 83 | SAINT1 | 20.40 | 18.87 | 27.10 | 20.40 | 18.87 | 27.10 | SAINT1 | 83 | ||||||
| 84 | FFASsuboptimal | 20.40 | 17.72 | 20.99 | 21.84 | 19.73 | 21.66 | FFASsuboptimal | 84 | ||||||
| 85 | A−TASSER | 20.40 | 17.72 | 27.21 | 43.68 | 35.63 | 43.09 | 1.76 | 1.52 | 1.44 | A−TASSER | 85 | |||
| 86 | igor | 20.40 | 17.15 | 28.14 | 20.40 | 17.15 | 28.14 | igor | 86 | ||||||
| 87 | POEMQA | 20.40 | 16.76 | 23.45 | 30.46 | 26.05 | 34.26 | 0.14 | 0.16 | 0.29 | POEMQA | 87 | |||
| 88 | StruPPi | 20.40 | 16.09 | 23.53 | 20.40 | 16.09 | 23.53 | StruPPi | 88 | ||||||
| 89 | MUFOLD−Server | 20.40 | 15.33 | 26.04 | 20.69 | 16.28 | 26.05 | MUFOLD−Server | 89 | ||||||
| 90 | EB_AMU_Physics | 20.40 | 15.04 | 23.31 | 20.40 | 17.91 | 23.31 | EB_AMU_Physics | 90 | ||||||
| 91 | HHpred5 | 20.11 | 18.01 | 21.48 | 20.11 | 18.01 | 21.48 | HHpred5 | 91 | ||||||
| 92 | mahmood−torda−server | 20.11 | 16.09 | 18.19 | 20.11 | 17.15 | 19.32 | mahmood−torda−server | 92 | ||||||
| 93 | 3D−JIGSAW_V3 | 20.11 | 14.75 | 20.10 | 20.69 | 16.95 | 20.43 | 3D−JIGSAW_V3 | 93 | ||||||
| 94 | 3D−JIGSAW_AEP | 19.83 | 17.05 | 19.96 | 19.83 | 17.15 | 19.99 | 3D−JIGSAW_AEP | 94 | ||||||
| 95 | DistillSN | 19.83 | 16.86 | 23.99 | 22.41 | 18.97 | 29.93 | DistillSN | 95 | ||||||
| 96 | HHpred4 | 19.83 | 14.46 | 22.60 | 19.83 | 14.46 | 22.60 | HHpred4 | 96 | ||||||
| 97 | RANDOM | 19.38 | 15.93 | 19.38 | 19.38 | 15.93 | 19.38 | RANDOM | 97 | ||||||
| 98 | 3Dpro | 19.25 | 17.53 | 25.54 | 21.55 | 19.44 | 25.54 | 3Dpro | 98 | ||||||
| 99 | 3DShot2 | 19.25 | 16.19 | 21.76 | 19.25 | 16.19 | 21.76 | 3DShot2 | 99 | ||||||
| 100 | COMA | 18.97 | 16.57 | 19.55 | 27.59 | 25.86 | 21.47 | 0.13 | COMA | 100 | |||||
| 101 | keasar−server | 18.97 | 16.09 | 18.24 | 18.97 | 16.09 | 19.29 | keasar−server | 101 | ||||||
| 102 | forecast | 18.68 | 17.24 | 23.23 | 20.98 | 17.53 | 24.19 | forecast | 102 | ||||||
| 103 | FUGUE_KM | 18.68 | 15.42 | 13.67 | 22.70 | 20.02 | 21.78 | FUGUE_KM | 103 | ||||||
| 104 | xianmingpan | 18.39 | 15.71 | 22.92 | 18.39 | 15.71 | 22.92 | xianmingpan | 104 | ||||||
| 105 | DelCLab | 18.39 | 14.56 | 20.01 | 19.83 | 18.30 | 20.88 | DelCLab | 105 | ||||||
| 106 | nFOLD3 | 18.39 | 14.18 | 21.71 | 48.85 | 42.91 | 46.89 | 2.39 | 2.56 | 1.94 | nFOLD3 | 106 | |||
| 107 | SAM−T06−server | 18.10 | 15.61 | 21.58 | 20.40 | 19.83 | 21.58 | SAM−T06−server | 107 | ||||||
| 108 | huber−torda−server | 18.10 | 15.23 | 14.86 | 21.26 | 18.58 | 19.38 | huber−torda−server | 108 | ||||||
| 109 | keasar | 18.10 | 15.04 | 17.10 | 40.80 | 36.40 | 41.30 | 1.41 | 1.63 | 1.21 | keasar | 109 | |||
| 110 | LevittGroup | 18.10 | 14.08 | 17.02 | 31.61 | 28.54 | 35.18 | 0.28 | 0.51 | 0.41 | LevittGroup | 110 | |||
| 111 | mGenTHREADER | 18.10 | 13.41 | 18.83 | 18.10 | 13.41 | 18.83 | mGenTHREADER | 111 | ||||||
| 112 | Jiang_Zhu | 17.53 | 15.42 | 21.90 | 29.02 | 24.23 | 32.99 | 0.13 | Jiang_Zhu | 112 | |||||
| 113 | LOOPP_Server | 17.53 | 12.74 | 20.13 | 45.40 | 39.66 | 44.56 | 1.97 | 2.10 | 1.63 | LOOPP_Server | 113 | |||
| 114 | Frankenstein | 17.53 | 12.36 | 18.00 | 19.25 | 15.80 | 25.57 | Frankenstein | 114 | ||||||
| 115 | SAM−T02−server | 17.24 | 16.09 | 15.30 | 20.40 | 19.83 | 15.30 | SAM−T02−server | 115 | ||||||
| 116 | IBT_LT | 17.24 | 13.98 | 17.05 | 17.24 | 13.98 | 17.05 | IBT_LT | 116 | ||||||
| 117 | panther_server | 16.95 | 15.80 | 10.62 | 17.24 | 15.80 | 12.18 | panther_server | 117 | ||||||
| 118 | Phragment | 16.95 | 15.52 | 21.39 | 20.69 | 19.83 | 25.47 | Phragment | 118 | ||||||
| 119 | COMA−M | 16.95 | 15.04 | 18.13 | 18.97 | 16.57 | 19.55 | COMA−M | 119 | ||||||
| 120 | SAMUDRALA | 16.67 | 15.90 | 20.72 | 17.24 | 15.90 | 20.72 | SAMUDRALA | 120 | ||||||
| 121 | schenk−torda−server | 16.67 | 15.42 | 19.00 | 18.10 | 15.71 | 22.23 | schenk−torda−server | 121 | ||||||
| 122 | OLGAFS | 16.67 | 15.13 | 10.01 | 16.67 | 15.52 | 10.01 | OLGAFS | 122 | ||||||
| 123 | Pcons_local | 16.67 | 14.65 | 14.92 | 16.67 | 14.65 | 14.92 | Pcons_local | 123 | ||||||
| 124 | Pcons_dot_net | 16.67 | 14.65 | 14.92 | 40.23 | 32.95 | 39.65 | 1.34 | 1.14 | 0.99 | Pcons_dot_net | 124 | |||
| 125 | Pcons_multi | 16.67 | 14.18 | 16.96 | 18.97 | 16.86 | 21.78 | Pcons_multi | 125 | ||||||
| 126 | FOLDpro | 16.38 | 14.85 | 17.38 | 21.84 | 20.69 | 27.59 | FOLDpro | 126 | ||||||
| 127 | pipe_int | 16.38 | 14.27 | 17.66 | 16.38 | 14.27 | 17.66 | pipe_int | 127 | ||||||
| 128 | FFASstandard | 16.38 | 13.89 | 14.24 | 16.38 | 13.89 | 14.44 | FFASstandard | 128 | ||||||
| 129 | FFASflextemplate | 16.38 | 13.89 | 14.24 | 16.38 | 13.89 | 14.44 | FFASflextemplate | 129 | ||||||
| 130 | TWPPLAB | 13.79 | 13.03 | 16.49 | 13.79 | 13.03 | 16.49 | TWPPLAB | 130 | ||||||
| 131 | Pushchino | 11.49 | 8.62 | 5.38 | 11.49 | 8.62 | 5.38 | Pushchino | 131 | ||||||
| 132 | rehtnap | 3.45 | 3.45 | 0.00 | 3.45 | 3.45 | 0.00 | rehtnap | 132 | ||||||
| 133 | Abagyan | Abagyan | 133 | ||||||||||||
| 134 | BHAGEERATH | BHAGEERATH | 134 | ||||||||||||
| 135 | CBSU | CBSU | 135 | ||||||||||||
| 136 | FEIG_REFINE | FEIG_REFINE | 136 | ||||||||||||
| 137 | Fiser−M4T | Fiser−M4T | 137 | ||||||||||||
| 138 | HCA | HCA | 138 | ||||||||||||
| 139 | LEE−SERVER | LEE−SERVER | 139 | ||||||||||||
| 140 | Linnolt−UH−CMB | Linnolt−UH−CMB | 140 | ||||||||||||
| 141 | NIM2 | NIM2 | 141 | ||||||||||||
| 142 | Nano_team | Nano_team | 142 | ||||||||||||
| 143 | NirBenTal | NirBenTal | 143 | ||||||||||||
| 144 | PHAISTOS | PHAISTOS | 144 | ||||||||||||
| 145 | PZ−UAM | PZ−UAM | 145 | ||||||||||||
| 146 | ProtAnG | ProtAnG | 146 | ||||||||||||
| 147 | RBO−Proteus | RBO−Proteus | 147 | ||||||||||||
| 148 | RPFM | RPFM | 148 | ||||||||||||
| 149 | ShakAbInitio | ShakAbInitio | 149 | ||||||||||||
| 150 | TsaiLab | TsaiLab | 150 | ||||||||||||
| 151 | UCDavisGenome | UCDavisGenome | 151 | ||||||||||||
| 152 | Wolfson−FOBIA | Wolfson−FOBIA | 152 | ||||||||||||
| 153 | YASARA | YASARA | 153 | ||||||||||||
| 154 | YASARARefine | YASARARefine | 154 | ||||||||||||
| 155 | Zhou−SPARKS | Zhou−SPARKS | 155 | ||||||||||||
| 156 | dill_ucsf | dill_ucsf | 156 | ||||||||||||
| 157 | dill_ucsf_extended | dill_ucsf_extended | 157 | ||||||||||||
| 158 | jacobson | jacobson | 158 | ||||||||||||
| 159 | mti | mti | 159 | ||||||||||||
| 160 | mumssp | mumssp | 160 | ||||||||||||
| 161 | psiphifoldings | psiphifoldings | 161 | ||||||||||||
| 162 | ricardo | ricardo | 162 | ||||||||||||
| 163 | rivilo | rivilo | 163 | ||||||||||||
| 164 | sessions | sessions | 164 | ||||||||||||
| 165 | test_http_server_01 | test_http_server_01 | 165 | ||||||||||||
| 166 | tripos_08 | tripos_08 | 166 | ||||||||||||