T0468
putative lipoprotein from Bacillus cereus
Target sequence:
>T0468 BcR103A, Bacillus cereus, 109 residues
MERASLNRIGKDVYYMQIKGEGTIEKVDGRNLRNYTLPAYDEDGVKKQITFRSTKKENDHKLNKYAFLRLYVDQDDNSKNEISSIEVKSYEEIQKADLPEKVKDKFTIK
Structure:
Method: NMR
Determined by: NESG
PDB ID: 2k5w
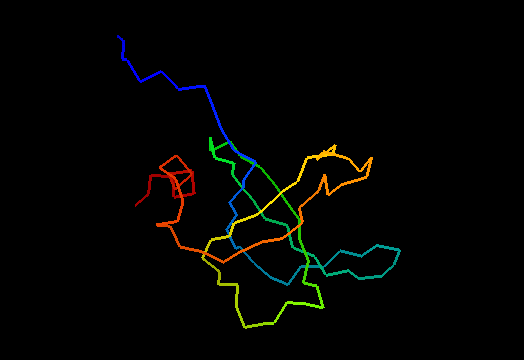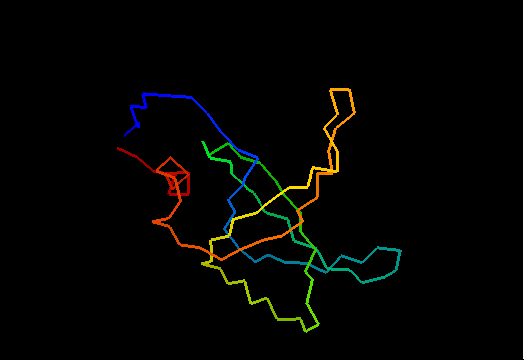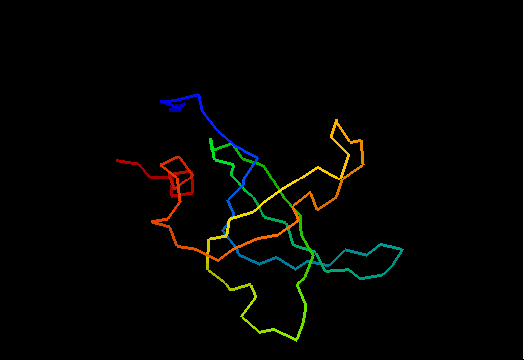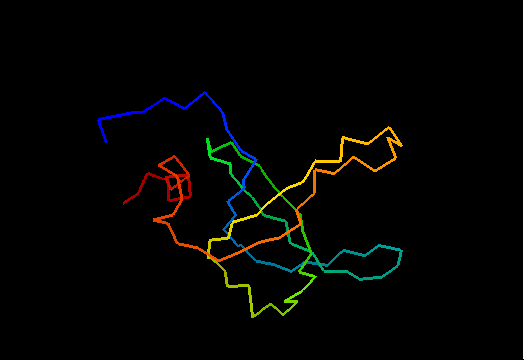
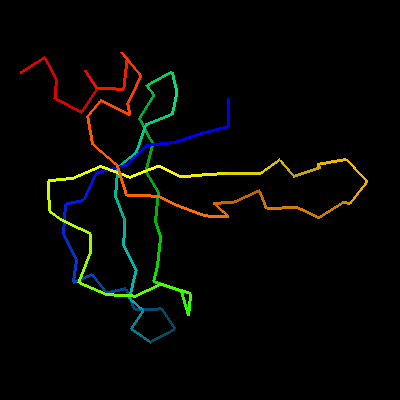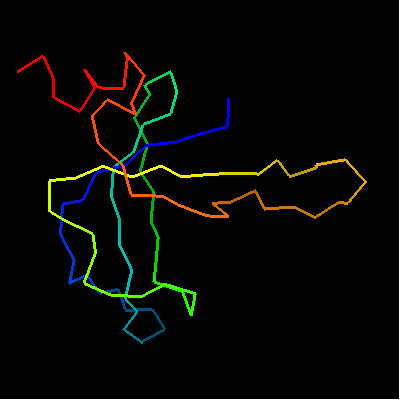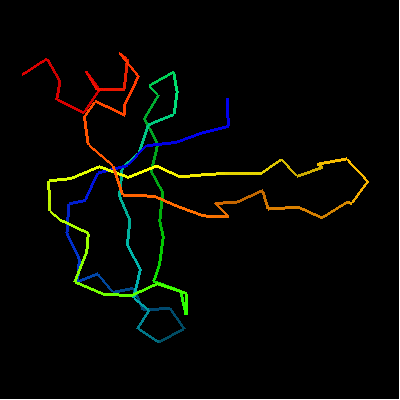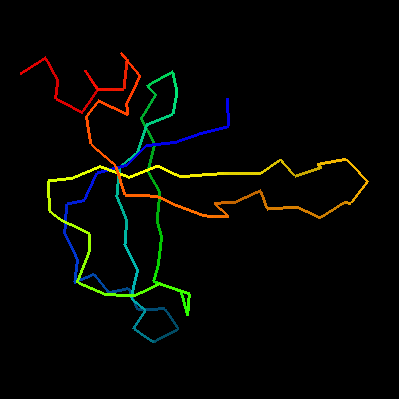
Domains: PyMOL of domains
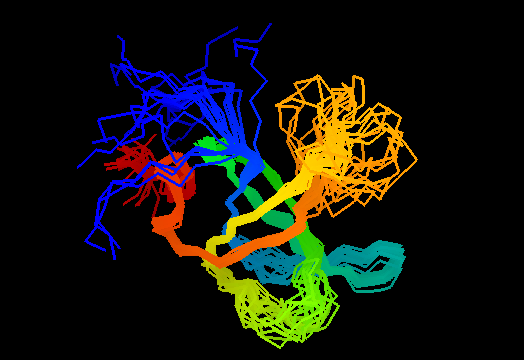
Single domain protein with poorly structured termini (mostly N-) and loops. Structurally variable regions were removed from NMR models for evaluation of predictions.
Structure classification:
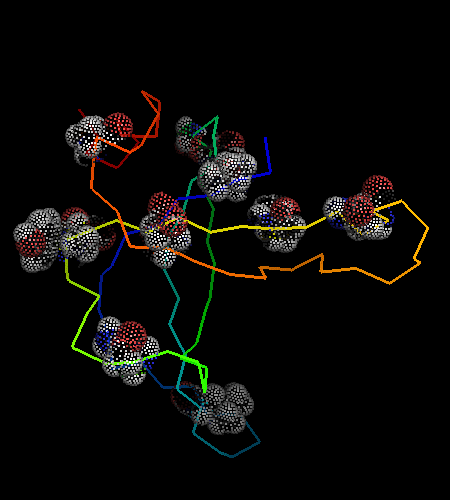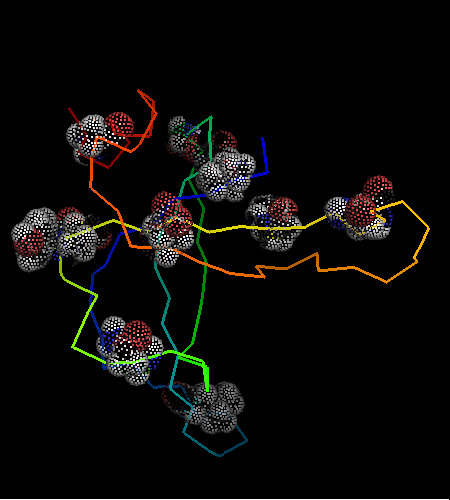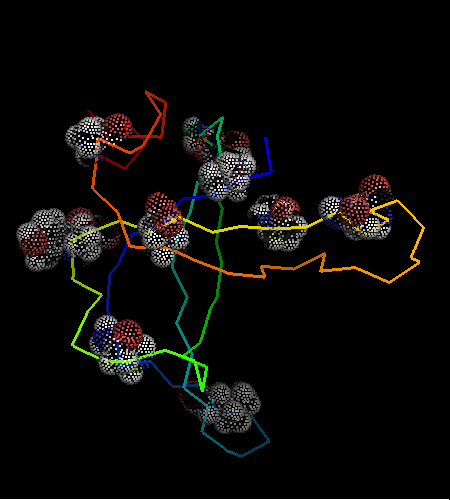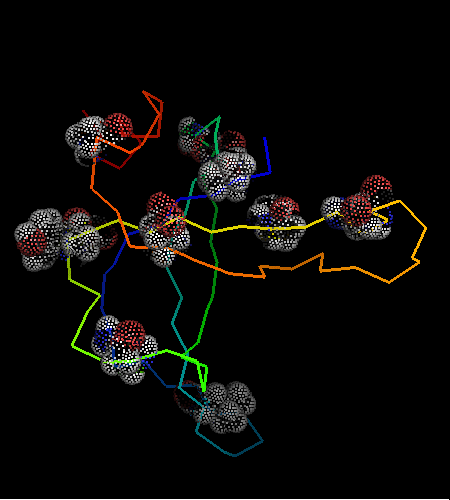
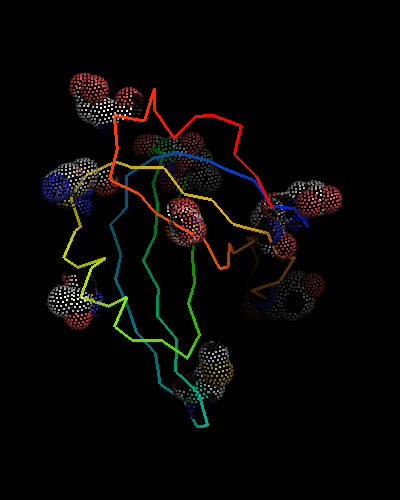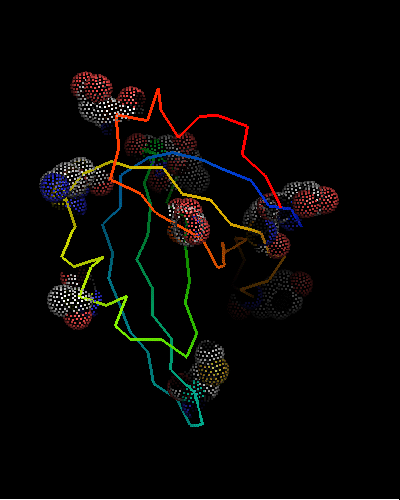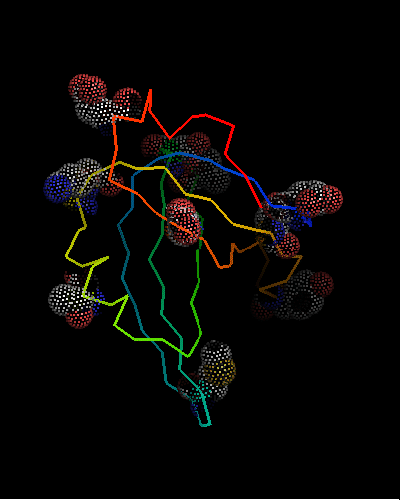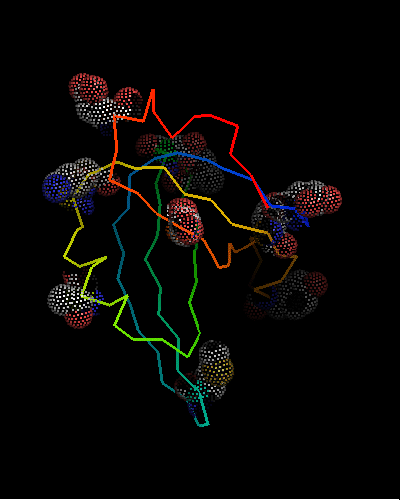
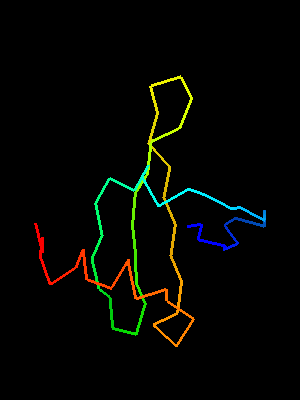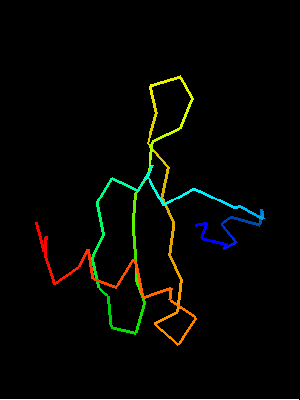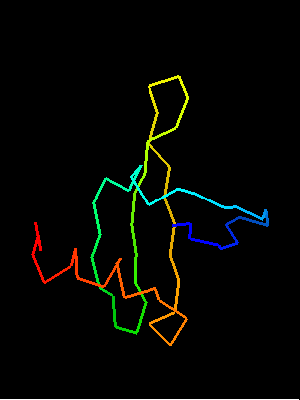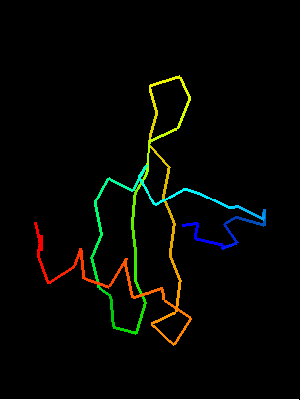
OB-fold. 5-stranded β-barrel, which in contrast to most OB-fold members is open between the 3rd and the 5th β-strands. The five strands are shown in the ribbon diagram below as dark blue, cyan, green, yellow and orange. In addition to the 5-stranded OB-core, this structure is elaborated with a long insertion between β-strands 1 and 2 housing a β-hairpin (which is an extension of the hairpin 1-2), and two C-terminal α-helices.
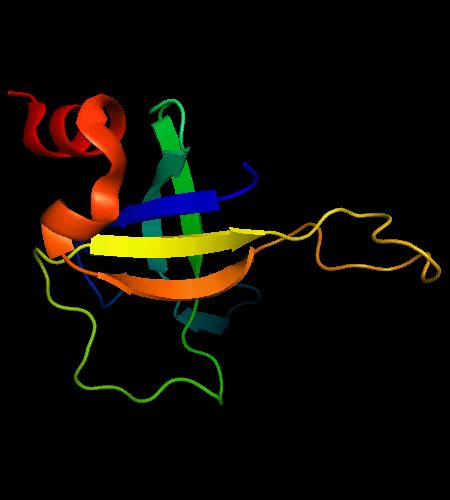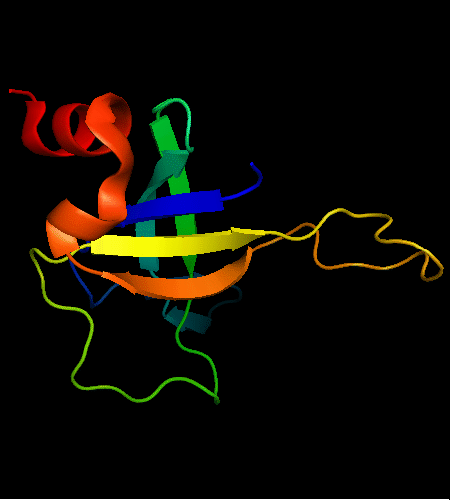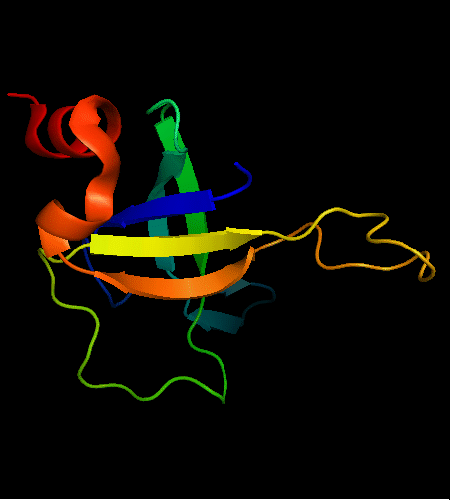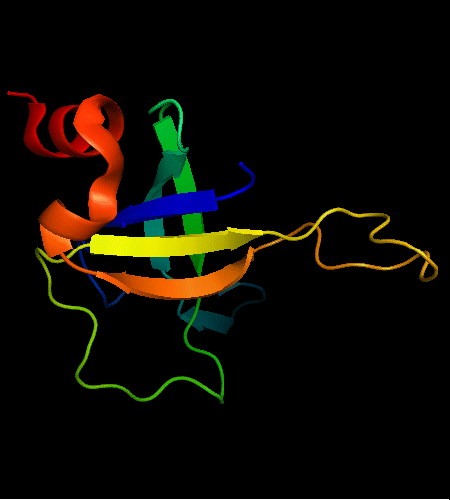
Cartoon diagram of T0468: 2k5w model 1 residues 11-110
CASP category:
Fold recognition. Fold similarity not detectable statistically by sequence methods known to us.
Closest templates:
The first COMPASS hit to OB-fold proteins is to the Nucleic acid - binding domain superfamily:
Subject = d1gm5a2 b.40.4.9 RecG "wedge" domain length=180 filtered_length=178 Neff=17.669 Smith-Waterman score = 41 Evalue = 4.14e+01 QUERY 5 SLNRIGKDVYYMQIKGEGTIEKVDGRNLRNYTLPAYDEDGVKKQITFRSTKKENDHKLNKYAFLRLY==VDQDDNskneISSIEVKSYEEIQKADLPEKVKDKF 106 CONSENSUS_1 5 QLNPIGGDEYYVQITGDGKKKGEKGDGGYSYTLKGYDEDGKEKEVTFTAGKKELDHNLRKGAYLKLY==VKGKKG....IKGIEVTSWEEVKKEDIPKKALEKL 106 +++ ++ +E ++ + ++++ ++ +G +++ D++G+ + V F+ + ++L + L++GA L ++ V+ ++G ++++++ ++E ++ E+ ++ L+ + CONSENSUS_2 53 PIAELRPGEKVTVAGIVVSVRVIRTKRGKMLIVTLEDGTGSLELVFFNGVYAKLKKLLKVGARLLVSGKVERFDG~~~~GLQLVHPDYELLDEEEALEGRLVPV 152 d1gm5a2 53 KLNDLLPGEKVTTQGKIVSVETKKFQNMNILTAVLSDGLVHVPLKWFNQDYLQTYLKQLTGKEVFVTGTVKSNAY~~~~TGQYEIHNAEVTPKEGEYVRRILPI 152
However, this is the hit #41 in the total list of hits and its E-value is around 41 (which indicates BTW how good COMPASS E-value estimate is), so it can hardy be called statistically significant. Moreover, proteins from all SCOP classes were found by COMPASS prior to this hit. Nevertheless, 9 consensus match residues from the above alignment were selected among 16 and shown as "dots" on the two OB-fold structures, map to structurally equivalent positions validating the COMPASS alignment:
According to structure, proteins from 'Nucleic acid - binding' superfamily are also most similar to T0468, and those include phage ssDNA - binding proteins with an open barrel.
Target sequence - PDB file inconsistencies:
NMR models contain two C-terminal residues 110-LE-111, not present in the target sequence. This regions was removed from NMR models due to disorder anyway.
T0468 2k5w.pdb T0468.pdb PyMOL PyMOL of domains
T0468 1 MERASLNRIGKDVYYMQIKGEGTIEKVDGRNLRNYTLPAYDEDGVKKQITFRSTKKENDHKLNKYAFLRLYVDQDDNSKNEISSIEVKSYEEIQKADLPEKVKDKFTIK-- 109 **********|||||||||||||||||||||||||||||||||||||||||||||****|||||||||||||||||********|||||||||||||||||||||||||~~ 2k5wA 1 MERASLNRIGKDVYYMQIKGEGTIEKVDGRNLRNYTLPAYDEDGVKKQITFRSTKKENDHKLNKYAFLRLYVDQDDNSKNEISSIEVKSYEEIQKADLPEKVKDKFTIKLE 111
Residue change log: remove 110-LE-111 as it is not present in target sequence; remove 1-MERASLNRIG-10, 56-KEND-59, 77-NSKNEISS-84 by 3.5Å SD cutoff;
Single domain protein: target 11-55, 60-76, 85-109 ; pdb 11-55, 60-76, 85-109
Sequence classification:
DUF1093 in Pfam, which includes just a few very close homologs. Below is PROMALS3D alignment of several most distant homologs detectable by PSI-BLAST. This alignment was not edited manually and may have errors. Biological sequences contain signal peptide (predicted as a helix at the N-terminus) not included in the target sequence.
Conservation: 9 7 9 5 7 6 5 5 555 9 6 5
T0468 1 M--ERA----------------------------------SLNR--IGKDVYYMQIKGEGTIEKVDG-----RNLRNYTLPAYDED---GVKKQITFRSTKKENDHKLNKYAFLRLYVDQDDNSKNEISSIEVKSYEEIQKADLPEKVKDKFTIK------------------------------------ 109T0467 1 M---------------------------------------DLNR--MGKDEYYVQITVDGKEVHSKADNGQKYKDYEYKLTGFDKD---GKEKELEFTAQ-----KNLRKEAFLRVYHSDK---------KGVSAWEEVKKDELPAKVKEKLGVK------------------------------------ 97gi_163762713 1 M--KKT--------IIILISTLVSLAVIAVVII---ATI-DFNR--VGKDHAYTLITENGDTEETVTGSGEVFTRYHYSQSAFTDE---GEEITVSFSSH-----KNLRHGAYLRLYMNH----------TDVTSYDEVQTEDVPAQALETLMQE------------------------------------ 121
gi_116334877 1 ----KG-----------------------------IGTIDRFDPF-LSYKVGYTKLPSEKEISPNNK------ATNGYWTAWFTDNTMGNEWTKLTLTTGL----NPGVKNPYVMAYHKG----------SYVKVARIVPRTYINQTYVSNLAALDALLNHKGNTATNNHDLKKYKTQIHISDTVQQLVTE 137gi_49398117 1 M--KRT--------LIIV--TIVLLIIILSLLLVRNEHLDRFNPL-LKEKVSYAKVPKDT--------------QDYRNITVYSKN---GEKKSYKLDFLG-----YDPTKEYVKVNHKG----------KYVRSIEYIT-KIFLAF-------------------------------------------- 101gi_116492027 1 M--RKG--------MIIS--GVVVVILLITF----GSAYFWYNSN-YGTQDYYTQIVDKGTKFVEKDDSNRSYVNYAYHQPAYNQA---SQKITVKFNGNL---ERPLKLHAYLKIGYNAKR--------NQVISWSKVDKKDVPKAALKQINP------------------------------------- 123gi_69249191 1 M--KK---------LIGV--LVVL---GIFL----GGSFAAYHYF-YGGEAYYTKITTNGEMSATQADNGEKFTTYTYKQAAYNKD---GEKKQVTLREER---EHPLKMNAYLKLKVNPR---------KGVLSWEEIKASEVPKNAAEKIDN------------------------------------- 118gi_116873589 1 MITKKR-------VLISL-----ATVLLAVCA-----IWEYAMPTDAAVKSYYLKVAEAGKPVKKN-----QFKGYEYTSKVYDDK---GKQKEIKFYSEK-----ELTKNEQFKVMIGEN---------KMVVNYKKIK--NVPDKIKYLAEK------------------------------------- 113gi_65320434 1 M--NKRRNDYEDFVLCIL--GIGLPIYLLPFIL--RGDFDRFNPI-AEEKNVYAIAKGYGVPDYH------HKGRAMYSLKSFDES---GSRNEYTVGTST---PNDFIRKTYLRIHVKG----------KYVYSYEAISEKDIPEKIRKQLEV------EIK---------------------------- 128Consensus_aa: M..c+.................................h..hp...h..cs.Yhbl...sp...pp......b....Yp..s@scp...tpp.phph........p....p.@l+l.hp...........p.V.shcblppp.ls..h.pph.......................................Consensus_ss: h hh hhhhhhhh eeeeeee ee eeeeeeeeee eeeeeee eeeeeee eeeehhh hhh hhhhhhh
Comments:
This protein is a close homolog of T0467:
PSIPRED ------------EEEEEE----EEE-----------EEEEEEEEE-----EEEEEEE--------------EEEEEEE------------EEEEHHH--HHH--HHHHHHH---
T0468 MERASLNRIGKDVYYMQIKGEGTIEKV-----DGRNLRNYTLPAYDEDGVKKQITFRSTKKENDHKLNKYAFLRLYVDQDDNSKNEISSIEVKSYEEIQKADLPEKVKDKFTIK
T0467 MD---LNRMGKDEYYVQITVDGKEVHSKADNGQKYKDYEYKLTGFDKDGKEKELEFTAQK-----NLRKEAFLRVYHSDK---------KGVSAWEEVKKDELPAKVKEKLGVK
PSIPRED ------------EEEEEEE---EEEE-------EEEEEEEEEEEE-----EEEEEEE--------------EEEEEE--------------EEEEEEE------HHHHHHH---
This OB-fold was predicted de-novo by ROSETTA, which correctly indicated an open barrel. This prediction was very suggestive of the correct structure, because ROSETTA is biased towards local β-strand pairing, and OB-fold has a crossing loop to form H-bonds between β-strands 1 and 4, not to mention barrel closure present in most OB-folds with H-bonds between β-strands 3 and 5. Metaserver bioinfo.pl fails to find similarity to other OB-fold proteins. Although some servers used OB-folds as templates, it is unclear how significant those predictions are, as the templates were closed barrels and many severs used SH3-fold templates as well. The bias towards SH3 fold is likely caused by the C-terminal region, which shows strong local conformational similarity to Sso7d Chromo-domain DNA-binding proteins.
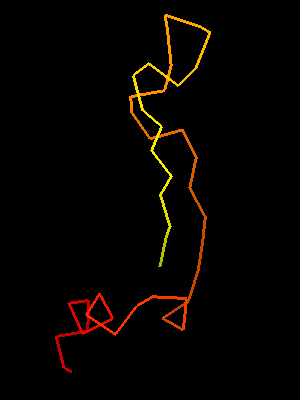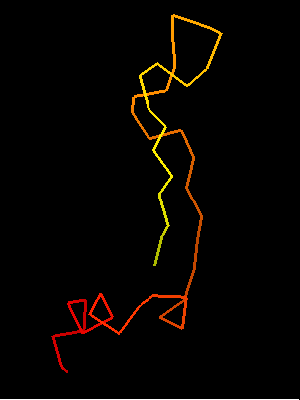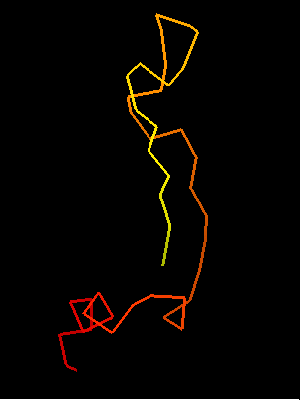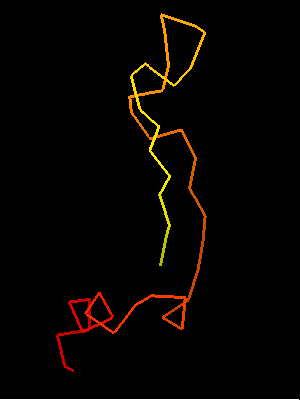
T0468 OB-fold C-terminal fragment:
2k5w model 1 residues 67-110
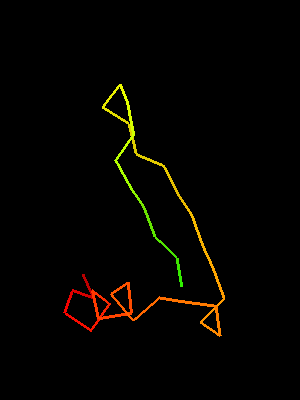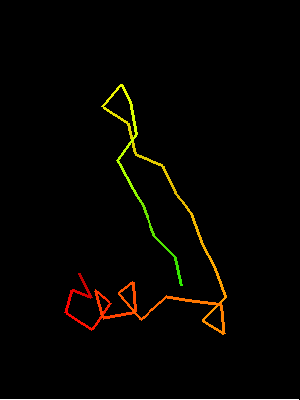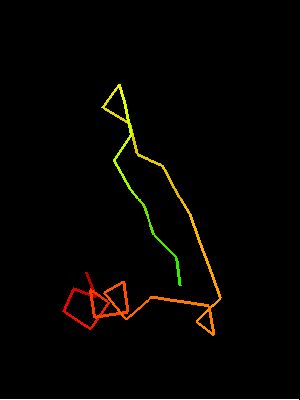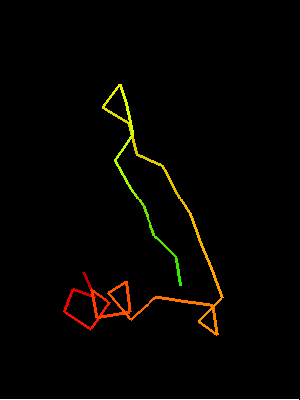
Sso7d SH3-fold C-terminal fragment:
2bf4 chain A residues 30-64
This similarity covers about 30 residues (half of Sso7d SH3-fold) and spans through a β-hairpin and two α-helices. The first helix is a single-turn helix characteristic of SH3 fold. Such a helix frequently structured as a 310-helix is present at this spatial location in the majority of SH3 fold proteins. Mutual orientations of the four secondary structural elements between the two fragments (from OB and SH3) is very similar, and sequence similarity reflects this.
2k5w 67 FLRLYVDQD[10]EVKSYEEIQKADLPEKVKDKFTI 107
2bf4_A 30 ISFTYDEGG----GKTGRGAVSEKDAPKELLQMLEK 61
EEEEEEEE EEEEEEEEHHHH HHHHHHHHHH
Therefore it is not surprising that servers find this similarity, in particular for T0467, which is a close homolog of T0468. However, upon closer inspection there are several positions with charged residues (highlighted red in the above alignment) aligned to hydrophobs (yellow). These positions have different exposure to solvent in the two structures and hint that the local similarity may not translate to global fold similarity. Indeed, the two C-terminal helices are essential core elements is SH3 structure of Sso7d, but are peripheral surface helices not contributing to the core in OB-fold of T0468. In addition, the surface of the hairpin buried in SH3 fold is exposed in T0468. These inconsistencies in hydrophobic patterns are very suggestive of global structural differences:
DALI-LITE does not detect similarity bewteen T0468 and Sso7d, but superimposes Sso7d with T0467, which is a close homolog of T0468. The central 3-stranded meander β-sheet in two proteins (shown in the back on the images above: blue-cyan-green on T0468 structure, while T0467 was superimposed, and green-yellow in Sso7d) is aligned, albeit with a very low Z-score of 0.3 (33 residues, RMSD 2Å). We agree that this is a globally meaningful alignment (only residues in capital letters are aligned), as it superimposes hydrophobic cores of both proteins.
DSSP ....lllllllleeeeeELLLllLEEHhhhllllllLEEEEEEEEEllllLEEEEEEEELL..LLLLlleeeeeelllllllleeeelllllllhhhhhllllllllllll
2k5q ....mdlnrmgkdeyyvQITVdgKEVHskadngqkyKDYEYKLTGFdkdgKEKELEFTAQK..NLRKeaflrvyhsdkkgvsaweevkkdelpakvkeklgvklehhhhhh
2bf4_A atvkfkykgeekevdisKIKK.vWRVG.........KMISFTYDEG....GGKTGRGAVSEkdAPKE..................................llqmlekqkk
DSSP leeeeeelleeeeeehhHEEE.eEEEL.........LEEEEEEELH....HHLEEEEEEELllLLHH..................................hhhhhhhlll
As a summary, superposition of locally similar fragments does not result in a global superpositions of structural cores of the two folds, and does not result in a reasonable fold prediction. Vice versa, global superposition of the cores leaves the two locally similar fragments as non-equivalent parts of the two proteins, as they occupy very different spatial locations and carry out different structural roles.
Global similarity between OB and SH3 folds has been noticed previously1), and explained in terms of very distant evolutionary relationship (homology). It is possible that the two folds shared a common ancestor and are indeed homologous over the 3-stranded curved meander sheet, although definitive evidence for this is lacking. The meaning of the local fragment similarity between T0468 and Sso7d-like chromo-domain OB-fold proteins is unclear. It is conceivable that these similar fragments originated independently and conformational resemblance between them is due to chance.
Server predictions:
T0468:pdb 11-109:seq 11-109:FR; alignment
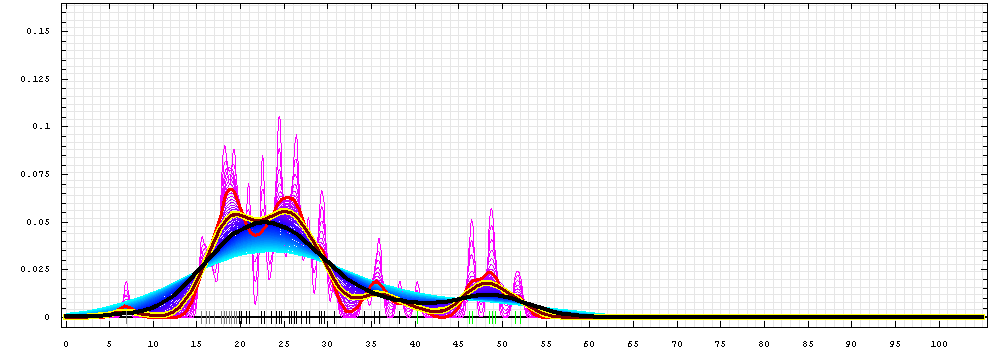
First models for T0468:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0468 | T0468 | T0468 | T0468 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | Pcons_dot_net | 52.01 | 42.62 | 55.55 | 2.03 | 2.06 | 1.60 | 52.01 | 42.62 | 55.55 | 1.67 | 1.66 | 1.28 | Pcons_dot_net | 1 |
| 2 | Pcons_multi | 51.44 | 39.56 | 59.97 | 1.97 | 1.67 | 2.03 | 51.44 | 43.30 | 59.97 | 1.61 | 1.75 | 1.72 | Pcons_multi | 2 |
| 3 | Chicken_George | 51.15 | 42.91 | 56.52 | 1.94 | 2.10 | 1.69 | 52.59 | 46.26 | 56.56 | 1.73 | 2.13 | 1.38 | Chicken_George | 3 |
| 4 | FAMS−multi | 50.00 | 41.00 | 56.40 | 1.83 | 1.85 | 1.68 | 50.58 | 41.95 | 58.74 | 1.53 | 1.57 | 1.60 | FAMS−multi | 4 |
| 5 | Jiang_Zhu | 50.00 | 38.31 | 47.96 | 1.83 | 1.51 | 0.85 | 50.00 | 38.31 | 48.48 | 1.47 | 1.10 | 0.57 | Jiang_Zhu | 5 |
| 6 | SAMUDRALA | 49.71 | 44.16 | 56.53 | 1.80 | 2.26 | 1.69 | 50.00 | 44.16 | 57.10 | 1.47 | 1.86 | 1.43 | SAMUDRALA | 6 |
| 7 | 3D−JIGSAW_V3 | 49.14 | 38.79 | 54.11 | 1.74 | 1.57 | 1.46 | 50.29 | 42.15 | 56.84 | 1.50 | 1.60 | 1.41 | 3D−JIGSAW_V3 | 7 |
| 8 | Sternberg | 49.14 | 38.79 | 54.11 | 1.74 | 1.57 | 1.46 | 49.14 | 40.90 | 54.11 | 1.38 | 1.43 | 1.13 | Sternberg | 8 |
| 9 | 3D−JIGSAW_AEP | 48.85 | 41.95 | 56.81 | 1.72 | 1.97 | 1.72 | 50.29 | 43.49 | 56.81 | 1.50 | 1.77 | 1.40 | 3D−JIGSAW_AEP | 9 |
| 10 | RAPTOR | 48.56 | 40.90 | 50.11 | 1.69 | 1.84 | 1.06 | 48.56 | 40.90 | 50.11 | 1.32 | 1.43 | 0.73 | RAPTOR | 10 |
| 11 | MUSTER | 48.56 | 37.64 | 51.31 | 1.69 | 1.43 | 1.18 | 48.56 | 37.64 | 51.31 | 1.32 | 1.01 | 0.85 | MUSTER | 11 |
| 12 | 3DShot1 | 48.56 | 34.58 | 52.35 | 1.69 | 1.04 | 1.28 | 48.56 | 34.58 | 52.35 | 1.32 | 0.61 | 0.95 | 3DShot1 | 12 |
| 13 | KudlatyPredHuman | 48.28 | 38.51 | 57.88 | 1.66 | 1.54 | 1.83 | 48.28 | 39.46 | 57.88 | 1.29 | 1.25 | 1.51 | KudlatyPredHuman | 13 |
| 14 | Hao_Kihara | 48.28 | 33.52 | 54.61 | 1.66 | 0.90 | 1.51 | 48.28 | 33.52 | 54.61 | 1.29 | 0.47 | 1.18 | Hao_Kihara | 14 |
| 15 | mufold | 47.99 | 39.27 | 55.89 | 1.63 | 1.63 | 1.63 | 47.99 | 39.27 | 55.89 | 1.27 | 1.22 | 1.31 | mufold | 15 |
| 16 | PRI−Yang−KiharA | 47.41 | 35.73 | 47.61 | 1.57 | 1.18 | 0.82 | 47.41 | 35.73 | 47.61 | 1.21 | 0.76 | 0.48 | PRI−Yang−KiharA | 16 |
| 17 | LevittGroup | 47.13 | 40.61 | 55.56 | 1.55 | 1.80 | 1.60 | 47.13 | 40.61 | 55.56 | 1.18 | 1.40 | 1.28 | LevittGroup | 17 |
| 18 | Zhang | 47.13 | 32.66 | 56.65 | 1.55 | 0.79 | 1.71 | 47.99 | 38.70 | 57.64 | 1.27 | 1.15 | 1.49 | Zhang | 18 |
| 19 | fams−ace2 | 46.55 | 38.12 | 55.75 | 1.49 | 1.49 | 1.62 | 52.01 | 40.13 | 59.89 | 1.67 | 1.33 | 1.71 | fams−ace2 | 19 |
| 20 | Zhang−Server | 46.55 | 36.97 | 54.93 | 1.49 | 1.34 | 1.54 | 46.55 | 36.97 | 54.93 | 1.12 | 0.92 | 1.21 | Zhang−Server | 20 |
| 21 | McGuffin | 46.55 | 36.97 | 54.93 | 1.49 | 1.34 | 1.54 | 48.56 | 37.64 | 54.93 | 1.32 | 1.01 | 1.21 | McGuffin | 21 |
| 22 | FAMSD | 46.55 | 35.82 | 47.64 | 1.49 | 1.19 | 0.82 | 46.55 | 35.82 | 48.90 | 1.12 | 0.77 | 0.61 | FAMSD | 22 |
| 23 | Jones−UCL | 46.26 | 38.98 | 52.50 | 1.46 | 1.60 | 1.30 | 46.26 | 38.98 | 52.50 | 1.09 | 1.18 | 0.97 | Jones−UCL | 23 |
| 24 | circle | 46.26 | 37.84 | 46.42 | 1.46 | 1.45 | 0.70 | 46.26 | 37.84 | 48.90 | 1.09 | 1.04 | 0.61 | circle | 24 |
| 25 | SAINT1 | 44.83 | 39.46 | 50.62 | 1.32 | 1.66 | 1.11 | 44.83 | 39.46 | 50.62 | 0.95 | 1.25 | 0.78 | SAINT1 | 25 |
| 26 | MULTICOM | 42.53 | 37.93 | 53.45 | 1.09 | 1.46 | 1.39 | 43.39 | 37.93 | 53.81 | 0.80 | 1.05 | 1.10 | MULTICOM | 26 |
| 27 | TASSER | 42.53 | 35.06 | 52.96 | 1.09 | 1.10 | 1.34 | 42.53 | 35.06 | 57.24 | 0.72 | 0.67 | 1.45 | TASSER | 27 |
| 28 | Zico | 40.80 | 36.40 | 50.10 | 0.92 | 1.27 | 1.06 | 46.84 | 37.26 | 55.27 | 1.15 | 0.96 | 1.25 | Zico | 28 |
| 29 | ZicoFullSTP | 40.80 | 36.40 | 50.10 | 0.92 | 1.27 | 1.06 | 47.99 | 37.84 | 55.19 | 1.27 | 1.04 | 1.24 | ZicoFullSTP | 29 |
| 30 | 3DShotMQ | 40.80 | 32.18 | 43.74 | 0.92 | 0.73 | 0.44 | 40.80 | 32.18 | 43.74 | 0.54 | 0.30 | 0.09 | 3DShotMQ | 30 |
| 31 | pro−sp3−TASSER | 40.23 | 36.21 | 48.90 | 0.87 | 1.24 | 0.95 | 48.28 | 39.46 | 56.91 | 1.29 | 1.25 | 1.41 | pro−sp3−TASSER | 31 |
| 32 | A−TASSER | 39.66 | 36.21 | 49.13 | 0.81 | 1.24 | 0.97 | 47.70 | 40.52 | 57.24 | 1.24 | 1.39 | 1.45 | A−TASSER | 32 |
| 33 | BioSerf | 38.22 | 31.13 | 43.28 | 0.67 | 0.60 | 0.40 | 38.22 | 31.13 | 46.91 | 0.28 | 0.16 | 0.41 | BioSerf | 33 |
| 34 | RBO−Proteus | 35.92 | 32.09 | 46.18 | 0.44 | 0.72 | 0.68 | 35.92 | 32.09 | 46.18 | 0.05 | 0.29 | 0.33 | RBO−Proteus | 34 |
| 35 | huber−torda−server | 35.92 | 28.83 | 32.93 | 0.44 | 0.30 | 35.92 | 28.83 | 33.74 | 0.05 | huber−torda−server | 35 | |||
| 36 | GS−KudlatyPred | 35.34 | 31.03 | 43.67 | 0.39 | 0.58 | 0.43 | 41.09 | 35.73 | 43.67 | 0.57 | 0.76 | 0.08 | GS−KudlatyPred | 36 |
| 37 | keasar | 35.34 | 29.60 | 49.00 | 0.39 | 0.40 | 0.96 | 35.34 | 31.80 | 49.54 | 0.25 | 0.67 | keasar | 37 | |
| 38 | POEM | 35.06 | 31.03 | 41.71 | 0.36 | 0.58 | 0.24 | 35.34 | 31.03 | 50.83 | 0.15 | 0.80 | POEM | 38 | |
| 39 | BAKER−ROBETTA | 34.20 | 30.56 | 50.04 | 0.27 | 0.52 | 1.06 | 34.20 | 30.56 | 50.04 | 0.09 | 0.72 | BAKER−ROBETTA | 39 | |
| 40 | TJ_Jiang | 31.03 | 24.14 | 37.93 | 31.03 | 27.49 | 38.54 | TJ_Jiang | 40 | ||||||
| 41 | FALCON | 30.75 | 27.87 | 39.91 | 0.18 | 0.07 | 31.32 | 28.07 | 40.45 | FALCON | 41 | ||||
| 42 | POEMQA | 30.17 | 25.57 | 40.62 | 0.13 | 34.77 | 31.13 | 50.83 | 0.16 | 0.80 | POEMQA | 42 | |||
| 43 | LEE | 30.17 | 23.47 | 38.93 | 30.17 | 25.57 | 47.05 | 0.42 | LEE | 43 | |||||
| 44 | ABIpro | 29.89 | 25.96 | 42.16 | 0.29 | 29.89 | 26.15 | 42.93 | 0.01 | ABIpro | 44 | ||||
| 45 | POISE | 29.60 | 27.59 | 43.42 | 0.14 | 0.41 | 29.60 | 27.59 | 45.10 | 0.23 | POISE | 45 | |||
| 46 | MUFOLD−MD | 29.60 | 27.49 | 43.15 | 0.13 | 0.38 | 35.63 | 30.65 | 43.15 | 0.02 | 0.10 | 0.03 | MUFOLD−MD | 46 | |
| 47 | SAM−T08−server | 29.60 | 25.19 | 37.97 | 29.60 | 25.19 | 38.80 | SAM−T08−server | 47 | ||||||
| 48 | MULTICOM−REFINE | 29.31 | 26.82 | 40.22 | 0.05 | 0.10 | 32.47 | 28.83 | 43.24 | 0.04 | MULTICOM−REFINE | 48 | |||
| 49 | Sasaki−Cetin−Sasai | 29.31 | 26.25 | 44.53 | 0.52 | 31.03 | 26.25 | 44.78 | 0.19 | Sasaki−Cetin−Sasai | 49 | ||||
| 50 | Elofsson | 29.02 | 25.77 | 39.96 | 0.07 | 29.89 | 26.82 | 40.29 | Elofsson | 50 | |||||
| 51 | MUProt | 29.02 | 25.57 | 40.31 | 0.10 | 32.47 | 28.26 | 43.29 | 0.04 | MUProt | 51 | ||||
| 52 | MULTICOM−CLUSTER | 29.02 | 25.38 | 40.00 | 0.07 | 37.93 | 33.14 | 43.58 | 0.26 | 0.42 | 0.07 | MULTICOM−CLUSTER | 52 | ||
| 53 | GeneSilico | 29.02 | 22.61 | 40.94 | 0.17 | 30.46 | 24.52 | 40.94 | GeneSilico | 53 | |||||
| 54 | Handl−Lovell | 28.45 | 25.77 | 34.42 | 35.63 | 32.76 | 40.44 | 0.02 | 0.37 | Handl−Lovell | 54 | ||||
| 55 | Bates_BMM | 28.45 | 24.33 | 37.47 | 47.13 | 40.42 | 49.86 | 1.18 | 1.37 | 0.70 | Bates_BMM | 55 | |||
| 56 | SMEG−CCP | 28.45 | 23.37 | 39.51 | 0.03 | 28.45 | 23.37 | 39.51 | SMEG−CCP | 56 | |||||
| 57 | SAM−T08−human | 28.16 | 23.28 | 40.30 | 0.10 | 34.48 | 28.35 | 48.63 | 0.58 | SAM−T08−human | 57 | ||||
| 58 | FLOUDAS | 28.16 | 23.18 | 31.89 | 43.10 | 35.06 | 46.85 | 0.77 | 0.67 | 0.40 | FLOUDAS | 58 | |||
| 59 | LOOPP_Server | 27.87 | 25.57 | 30.64 | 44.25 | 37.74 | 46.91 | 0.89 | 1.02 | 0.41 | LOOPP_Server | 59 | |||
| 60 | HHpred5 | 27.87 | 22.51 | 37.91 | 27.87 | 22.51 | 37.91 | HHpred5 | 60 | ||||||
| 61 | Ozkan−Shell | 27.59 | 24.52 | 38.13 | 28.74 | 26.63 | 38.27 | Ozkan−Shell | 61 | ||||||
| 62 | DBAKER | 27.59 | 23.56 | 28.20 | 59.48 | 53.54 | 70.89 | 2.42 | 3.08 | 2.82 | DBAKER | 62 | |||
| 63 | Bilab−UT | 27.59 | 22.80 | 39.71 | 0.05 | 28.74 | 26.63 | 41.26 | Bilab−UT | 63 | |||||
| 64 | METATASSER | 27.01 | 22.03 | 48.06 | 0.86 | 47.99 | 41.48 | 55.02 | 1.27 | 1.51 | 1.22 | METATASSER | 64 | ||
| 65 | ZicoFullSTPFullData | 26.72 | 22.61 | 47.73 | 0.83 | 47.99 | 37.84 | 55.19 | 1.27 | 1.04 | 1.24 | ZicoFullSTPFullData | 65 | ||
| 66 | MULTICOM−CMFR | 26.44 | 23.18 | 34.90 | 26.44 | 23.18 | 34.90 | MULTICOM−CMFR | 66 | ||||||
| 67 | mariner1 | 26.44 | 22.22 | 32.93 | 28.74 | 25.48 | 36.54 | mariner1 | 67 | ||||||
| 68 | FEIG | 26.44 | 20.69 | 35.66 | 32.18 | 24.14 | 42.56 | FEIG | 68 | ||||||
| 69 | FFASsuboptimal | 26.44 | 19.83 | 26.90 | 26.44 | 21.07 | 30.65 | FFASsuboptimal | 69 | ||||||
| 70 | Phragment | 26.15 | 22.41 | 39.67 | 0.04 | 26.15 | 23.66 | 40.19 | Phragment | 70 | |||||
| 71 | Distill | 25.86 | 22.61 | 39.20 | 25.86 | 22.61 | 39.20 | Distill | 71 | ||||||
| 72 | PS2−server | 25.57 | 23.47 | 33.20 | 39.66 | 35.44 | 41.48 | 0.43 | 0.72 | PS2−server | 72 | ||||
| 73 | Scheraga | 25.57 | 22.41 | 33.67 | 27.30 | 22.41 | 33.67 | Scheraga | 73 | ||||||
| 74 | MULTICOM−RANK | 25.57 | 21.84 | 31.48 | 27.30 | 22.22 | 38.84 | MULTICOM−RANK | 74 | ||||||
| 75 | SHORTLE | 25.29 | 22.32 | 33.25 | 25.29 | 24.14 | 34.68 | SHORTLE | 75 | ||||||
| 76 | Kolinski | 25.29 | 21.46 | 35.53 | 34.20 | 26.15 | 41.65 | Kolinski | 76 | ||||||
| 77 | ProteinShop | 25.00 | 23.47 | 33.34 | 25.57 | 23.47 | 37.45 | ProteinShop | 77 | ||||||
| 78 | fleil | 25.00 | 22.32 | 27.79 | 25.29 | 22.41 | 28.14 | fleil | 78 | ||||||
| 79 | MidwayFolding | 24.71 | 21.36 | 32.26 | 48.56 | 40.90 | 50.11 | 1.32 | 1.43 | 0.73 | MidwayFolding | 79 | |||
| 80 | HHpred4 | 24.71 | 20.69 | 34.12 | 24.71 | 20.69 | 34.12 | HHpred4 | 80 | ||||||
| 81 | FFASflextemplate | 24.71 | 20.11 | 22.91 | 24.71 | 20.11 | 22.91 | FFASflextemplate | 81 | ||||||
| 82 | Phyre_de_novo | 24.43 | 22.51 | 38.25 | 30.75 | 25.10 | 39.76 | Phyre_de_novo | 82 | ||||||
| 83 | keasar−server | 24.43 | 20.21 | 32.12 | 24.43 | 21.46 | 32.12 | keasar−server | 83 | ||||||
| 84 | FFASstandard | 24.43 | 18.10 | 22.49 | 24.71 | 20.11 | 22.91 | FFASstandard | 84 | ||||||
| 85 | Phyre2 | 24.14 | 21.93 | 32.89 | 33.05 | 27.30 | 41.92 | Phyre2 | 85 | ||||||
| 86 | mGenTHREADER | 24.14 | 21.07 | 19.08 | 24.14 | 21.07 | 19.08 | mGenTHREADER | 86 | ||||||
| 87 | Wolynes | 23.56 | 20.79 | 36.97 | 24.71 | 22.80 | 40.02 | Wolynes | 87 | ||||||
| 88 | nFOLD3 | 23.56 | 20.11 | 30.41 | 25.57 | 23.28 | 32.01 | nFOLD3 | 88 | ||||||
| 89 | EB_AMU_Physics | 22.99 | 20.88 | 33.97 | 22.99 | 20.88 | 33.97 | EB_AMU_Physics | 89 | ||||||
| 90 | Softberry | 22.70 | 21.55 | 30.46 | 22.70 | 21.55 | 30.46 | Softberry | 90 | ||||||
| 91 | taylor | 22.70 | 21.46 | 29.20 | 26.15 | 21.46 | 31.98 | taylor | 91 | ||||||
| 92 | COMA | 22.70 | 19.25 | 28.25 | 22.70 | 20.40 | 30.70 | COMA | 92 | ||||||
| 93 | ACOMPMOD | 22.70 | 17.34 | 21.96 | 23.56 | 21.65 | 32.97 | ACOMPMOD | 93 | ||||||
| 94 | HHpred2 | 22.41 | 20.88 | 32.24 | 22.41 | 20.88 | 32.24 | HHpred2 | 94 | ||||||
| 95 | COMA−M | 22.41 | 20.31 | 20.28 | 22.70 | 20.31 | 29.18 | COMA−M | 95 | ||||||
| 96 | 3Dpro | 22.41 | 18.58 | 28.33 | 25.57 | 22.32 | 31.96 | 3Dpro | 96 | ||||||
| 97 | MeilerLabRene | 22.13 | 19.83 | 30.50 | 24.43 | 21.55 | 32.59 | MeilerLabRene | 97 | ||||||
| 98 | DistillSN | 21.55 | 17.53 | 31.34 | 23.56 | 20.69 | 31.34 | DistillSN | 98 | ||||||
| 99 | FALCON_CONSENSUS | 20.98 | 20.02 | 31.27 | 31.32 | 28.07 | 40.45 | FALCON_CONSENSUS | 99 | ||||||
| 100 | fais−server | 20.98 | 19.35 | 36.81 | 34.20 | 29.98 | 40.86 | 0.01 | fais−server | 100 | |||||
| 101 | StruPPi | 20.98 | 17.72 | 30.78 | 20.98 | 17.72 | 30.78 | StruPPi | 101 | ||||||
| 102 | Poing | 20.98 | 16.57 | 29.99 | 27.59 | 23.56 | 35.96 | Poing | 102 | ||||||
| 103 | FOLDpro | 20.69 | 19.54 | 22.48 | 21.84 | 19.73 | 25.96 | FOLDpro | 103 | ||||||
| 104 | PS2−manual | 20.69 | 18.58 | 32.84 | 39.66 | 35.44 | 41.48 | 0.43 | 0.72 | PS2−manual | 104 | ||||
| 105 | igor | 20.11 | 18.01 | 26.70 | 20.11 | 18.01 | 26.70 | igor | 105 | ||||||
| 106 | FUGUE_KM | 20.11 | 17.15 | 26.99 | 30.46 | 27.20 | 30.58 | FUGUE_KM | 106 | ||||||
| 107 | FrankensteinLong | 19.83 | 16.57 | 28.82 | 47.41 | 37.64 | 54.67 | 1.21 | 1.01 | 1.19 | FrankensteinLong | 107 | |||
| 108 | 3DShot2 | 19.83 | 16.38 | 25.70 | 19.83 | 16.38 | 25.70 | 3DShot2 | 108 | ||||||
| 109 | SAM−T06−server | 19.54 | 18.58 | 25.41 | 34.48 | 28.74 | 30.30 | SAM−T06−server | 109 | ||||||
| 110 | RANDOM | 19.25 | 16.91 | 19.25 | 19.25 | 16.91 | 19.25 | RANDOM | 110 | ||||||
| 111 | CpHModels | 19.25 | 15.61 | 27.39 | 19.25 | 15.61 | 27.39 | CpHModels | 111 | ||||||
| 112 | PSI | 19.25 | 13.89 | 30.29 | 32.18 | 28.35 | 48.15 | 0.53 | PSI | 112 | |||||
| 113 | MUFOLD−Server | 18.97 | 16.48 | 26.09 | 29.31 | 24.52 | 38.94 | MUFOLD−Server | 113 | ||||||
| 114 | schenk−torda−server | 18.68 | 15.42 | 20.99 | 19.25 | 18.49 | 24.23 | schenk−torda−server | 114 | ||||||
| 115 | IBT_LT | 18.39 | 16.09 | 20.56 | 18.39 | 16.09 | 20.56 | IBT_LT | 115 | ||||||
| 116 | forecast | 18.39 | 14.85 | 24.68 | 22.99 | 20.21 | 25.37 | forecast | 116 | ||||||
| 117 | mahmood−torda−server | 18.39 | 14.65 | 18.77 | 18.39 | 16.00 | 24.52 | mahmood−torda−server | 117 | ||||||
| 118 | GS−MetaServer2 | 18.10 | 14.94 | 10.62 | 19.54 | 16.09 | 10.65 | GS−MetaServer2 | 118 | ||||||
| 119 | GeneSilicoMetaServer | 18.10 | 14.94 | 10.62 | 19.54 | 16.09 | 10.65 | GeneSilicoMetaServer | 119 | ||||||
| 120 | pipe_int | 17.82 | 15.71 | 24.24 | 17.82 | 15.71 | 24.24 | pipe_int | 120 | ||||||
| 121 | Frankenstein | 17.82 | 15.52 | 24.63 | 24.71 | 21.65 | 32.27 | Frankenstein | 121 | ||||||
| 122 | Pushchino | 16.95 | 14.46 | 10.04 | 16.95 | 14.46 | 10.04 | Pushchino | 122 | ||||||
| 123 | OLGAFS | 16.38 | 16.09 | 15.25 | 19.83 | 17.34 | 19.66 | OLGAFS | 123 | ||||||
| 124 | TWPPLAB | 16.38 | 12.84 | 26.48 | 16.38 | 12.84 | 26.48 | TWPPLAB | 124 | ||||||
| 125 | Pcons_local | 16.09 | 14.37 | 24.09 | 25.00 | 23.28 | 24.09 | Pcons_local | 125 | ||||||
| 126 | DelCLab | 16.09 | 14.27 | 22.19 | 22.41 | 20.50 | 23.64 | DelCLab | 126 | ||||||
| 127 | panther_server | 15.52 | 13.89 | 22.08 | 15.52 | 13.89 | 22.08 | panther_server | 127 | ||||||
| 128 | SAM−T02−server | 15.52 | 11.69 | 17.20 | 22.99 | 17.82 | 26.16 | SAM−T02−server | 128 | ||||||
| 129 | rehtnap | 6.90 | 4.79 | 0.00 | 6.90 | 5.27 | 0.00 | rehtnap | 129 | ||||||
| 130 | AMU−Biology | AMU−Biology | 130 | ||||||||||||
| 131 | Abagyan | Abagyan | 131 | ||||||||||||
| 132 | BHAGEERATH | BHAGEERATH | 132 | ||||||||||||
| 133 | CBSU | CBSU | 133 | ||||||||||||
| 134 | FEIG_REFINE | FEIG_REFINE | 134 | ||||||||||||
| 135 | Fiser−M4T | Fiser−M4T | 135 | ||||||||||||
| 136 | HCA | HCA | 136 | ||||||||||||
| 137 | JIVE08 | JIVE08 | 137 | ||||||||||||
| 138 | LEE−SERVER | LEE−SERVER | 138 | ||||||||||||
| 139 | Linnolt−UH−CMB | Linnolt−UH−CMB | 139 | ||||||||||||
| 140 | NIM2 | NIM2 | 140 | ||||||||||||
| 141 | Nano_team | Nano_team | 141 | ||||||||||||
| 142 | NirBenTal | NirBenTal | 142 | ||||||||||||
| 143 | PHAISTOS | PHAISTOS | 143 | ||||||||||||
| 144 | PZ−UAM | PZ−UAM | 144 | ||||||||||||
| 145 | ProtAnG | ProtAnG | 145 | ||||||||||||
| 146 | RPFM | RPFM | 146 | ||||||||||||
| 147 | ShakAbInitio | ShakAbInitio | 147 | ||||||||||||
| 148 | TsaiLab | TsaiLab | 148 | ||||||||||||
| 149 | UCDavisGenome | UCDavisGenome | 149 | ||||||||||||
| 150 | Wolfson−FOBIA | Wolfson−FOBIA | 150 | ||||||||||||
| 151 | YASARA | YASARA | 151 | ||||||||||||
| 152 | YASARARefine | YASARARefine | 152 | ||||||||||||
| 153 | Zhou−SPARKS | Zhou−SPARKS | 153 | ||||||||||||
| 154 | dill_ucsf | dill_ucsf | 154 | ||||||||||||
| 155 | dill_ucsf_extended | dill_ucsf_extended | 155 | ||||||||||||
| 156 | fais@hgc | fais@hgc | 156 | ||||||||||||
| 157 | jacobson | jacobson | 157 | ||||||||||||
| 158 | mti | mti | 158 | ||||||||||||
| 159 | mumssp | mumssp | 159 | ||||||||||||
| 160 | psiphifoldings | psiphifoldings | 160 | ||||||||||||
| 161 | ricardo | ricardo | 161 | ||||||||||||
| 162 | rivilo | rivilo | 162 | ||||||||||||
| 163 | sessions | sessions | 163 | ||||||||||||
| 164 | test_http_server_01 | test_http_server_01 | 164 | ||||||||||||
| 165 | tripos_08 | tripos_08 | 165 | ||||||||||||
| 166 | xianmingpan | xianmingpan | 166 | ||||||||||||