GO annotation search and functional categorization
Among 18242 Diaphorina citri psyllid proteins, 8594 are assigned at least on GO term. Predicted GO terms with confidence scores greater than 10 points, 8 points and 6 points are considered to be 'very confident', 'confident' and 'probable', respectively. We further mapped the predicted GO terms to Generic GO slim, which presents a general categorization of GO content and is useful for the broad protein function classification.
The functional categorization listing is shown below. Clicking on the number of GO annotation count navigates to the page that shows the list of predicted GO terms associated with corresponding GO slim term
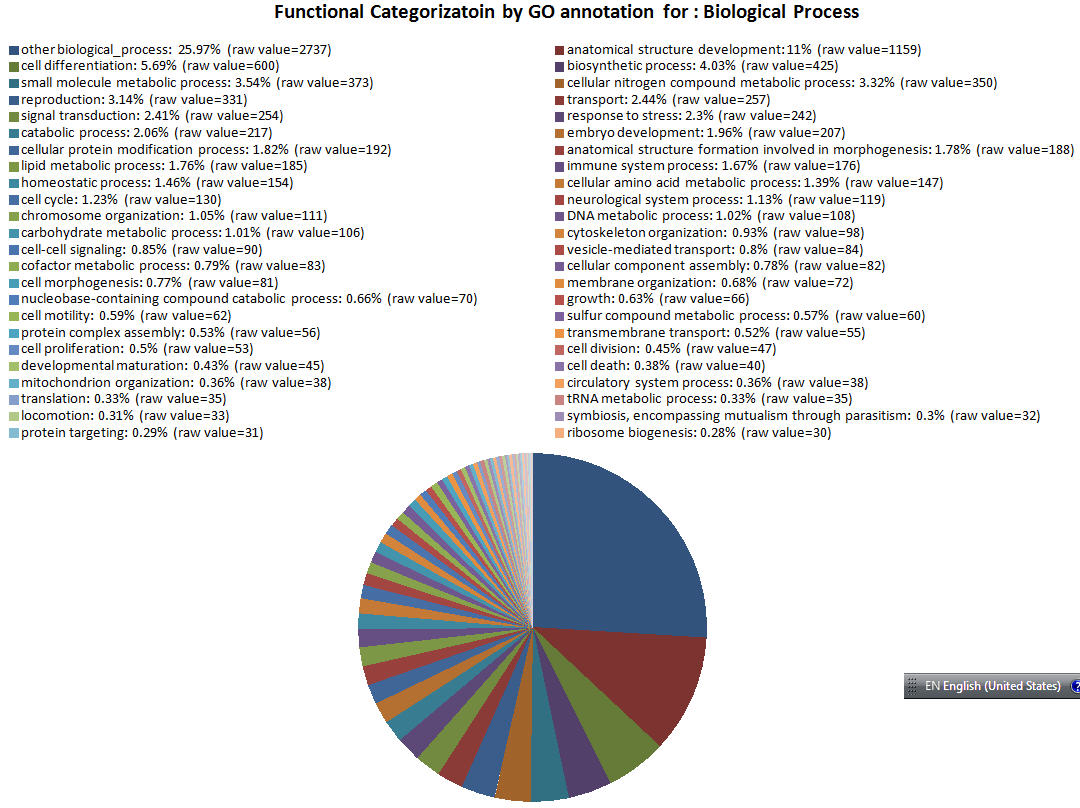
| GOID Link to GO |
Functional Category | GO Annotation Count to see GO terms |
|---|---|---|
| GO:0008150 | other biological_process | 2737 |
| GO:0048856 | anatomical structure development | 1159 |
| GO:0030154 | cell differentiation | 600 |
| GO:0009058 | biosynthetic process | 425 |
| GO:0044281 | small molecule metabolic process | 373 |
| GO:0034641 | cellular nitrogen compound metabolic process | 350 |
| GO:0000003 | reproduction | 331 |
| GO:0006810 | transport | 257 |
| GO:0007165 | signal transduction | 254 |
| GO:0006950 | response to stress | 242 |
| GO:0009056 | catabolic process | 217 |
| GO:0009790 | embryo development | 207 |
| GO:0006464 | cellular protein modification process | 192 |
| GO:0048646 | anatomical structure formation involved in morphogenesis | 188 |
| GO:0006629 | lipid metabolic process | 185 |
| GO:0002376 | immune system process | 176 |
| GO:0042592 | homeostatic process | 154 |
| GO:0006520 | cellular amino acid metabolic process | 147 |
| GO:0007049 | cell cycle | 130 |
| GO:0050877 | neurological system process | 119 |
| GO:0051276 | chromosome organization | 111 |
| GO:0006259 | DNA metabolic process | 108 |
| GO:0005975 | carbohydrate metabolic process | 106 |
| GO:0007010 | cytoskeleton organization | 98 |
| GO:0007267 | cell-cell signaling | 90 |
| GO:0016192 | vesicle-mediated transport | 84 |
| GO:0051186 | cofactor metabolic process | 83 |
| GO:0022607 | cellular component assembly | 82 |
| GO:0000902 | cell morphogenesis | 81 |
| GO:0061024 | membrane organization | 72 |
| GO:0034655 | nucleobase-containing compound catabolic process | 70 |
| GO:0040007 | growth | 66 |
| GO:0048870 | cell motility | 62 |
| GO:0006790 | sulfur compound metabolic process | 60 |
| GO:0006461 | protein complex assembly | 56 |
| GO:0055085 | transmembrane transport | 55 |
| GO:0008283 | cell proliferation | 53 |
| GO:0051301 | cell division | 47 |
| GO:0021700 | developmental maturation | 45 |
| GO:0008219 | cell death | 40 |
| GO:0007005 | mitochondrion organization | 38 |
| GO:0003013 | circulatory system process | 38 |
| GO:0006412 | translation | 35 |
| GO:0006399 | tRNA metabolic process | 35 |
| GO:0040011 | locomotion | 33 |
| GO:0044403 | symbiosis, encompassing mutualism through parasitism | 32 |
| GO:0006605 | protein targeting | 31 |
| GO:0042254 | ribosome biogenesis | 30 |
| GO:0019748 | secondary metabolic process | 30 |
| GO:0006091 | generation of precursor metabolites and energy | 27 |
| GO:0006913 | nucleocytoplasmic transport | 24 |
| GO:0051604 | protein maturation | 24 |
| GO:0007155 | cell adhesion | 23 |
| GO:0022618 | ribonucleoprotein complex assembly | 23 |
| GO:0006397 | mRNA processing | 22 |
| GO:0043473 | pigmentation | 22 |
| GO:0007059 | chromosome segregation | 20 |
| GO:0071554 | cell wall organization or biogenesis | 19 |
| GO:0007067 | mitosis | 18 |
| GO:0034330 | cell junction organization | 16 |
| GO:0065003 | macromolecular complex assembly | 15 |
| GO:0030705 | cytoskeleton-dependent intracellular transport | 15 |
| GO:0007568 | aging | 13 |
| GO:0006457 | protein folding | 11 |
| GO:0030198 | extracellular matrix organization | 11 |
| GO:0007034 | vacuolar transport | 10 |
| GO:0007009 | plasma membrane organization | 8 |
| GO:0071941 | nitrogen cycle metabolic process | 3 |
| GO:0015979 | photosynthesis | 2 |
| GO:0032196 | transposition | 1 |
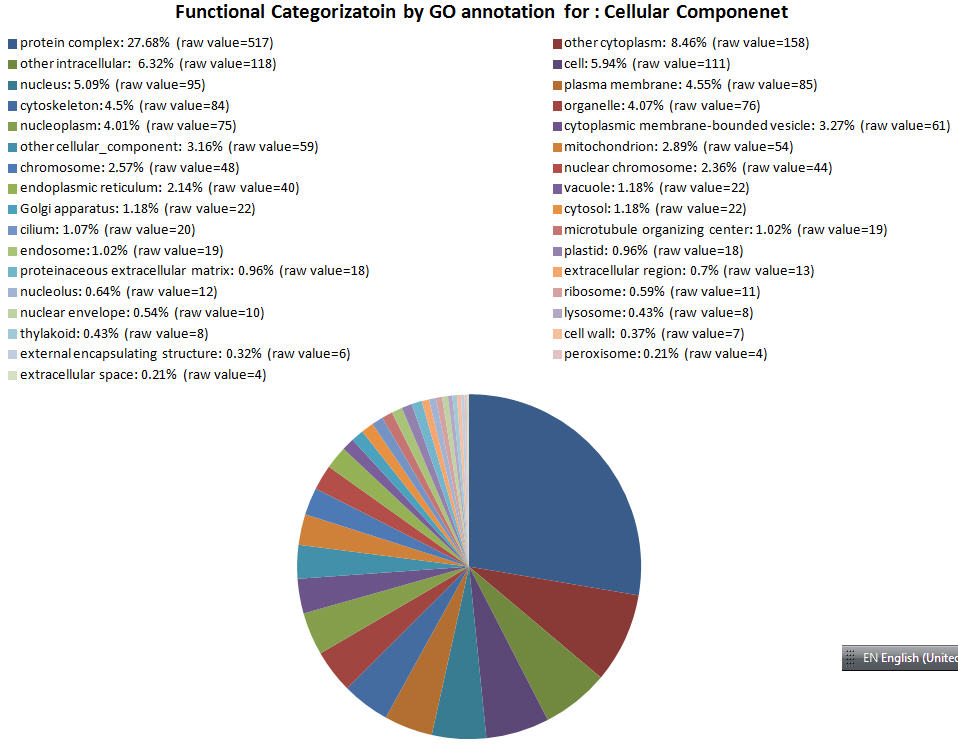
| GOID Link to GO |
Functional Category | GO Annotation Count to see GO terms |
|---|---|---|
| GO:0043234 | protein complex | 517 |
| GO:0005737 | other cytoplasm | 158 |
| GO:0005622 | other intracellular | 118 |
| GO:0005623 | cell | 111 |
| GO:0005634 | nucleus | 95 |
| GO:0005886 | plasma membrane | 85 |
| GO:0005856 | cytoskeleton | 84 |
| GO:0043226 | organelle | 76 |
| GO:0005654 | nucleoplasm | 75 |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | 61 |
| GO:0005575 | other cellular_component | 59 |
| GO:0005739 | mitochondrion | 54 |
| GO:0005694 | chromosome | 48 |
| GO:0000228 | nuclear chromosome | 44 |
| GO:0005783 | endoplasmic reticulum | 40 |
| GO:0005773 | vacuole | 22 |
| GO:0005794 | Golgi apparatus | 22 |
| GO:0005829 | cytosol | 22 |
| GO:0005929 | cilium | 20 |
| GO:0005815 | microtubule organizing center | 19 |
| GO:0005768 | endosome | 19 |
| GO:0009536 | plastid | 18 |
| GO:0005578 | proteinaceous extracellular matrix | 18 |
| GO:0005576 | extracellular region | 13 |
| GO:0005730 | nucleolus | 12 |
| GO:0005840 | ribosome | 11 |
| GO:0005635 | nuclear envelope | 10 |
| GO:0005764 | lysosome | 8 |
| GO:0009579 | thylakoid | 8 |
| GO:0005618 | cell wall | 7 |
| GO:0030312 | external encapsulating structure | 6 |
| GO:0005777 | peroxisome | 4 |
| GO:0005615 | extracellular space | 4 |
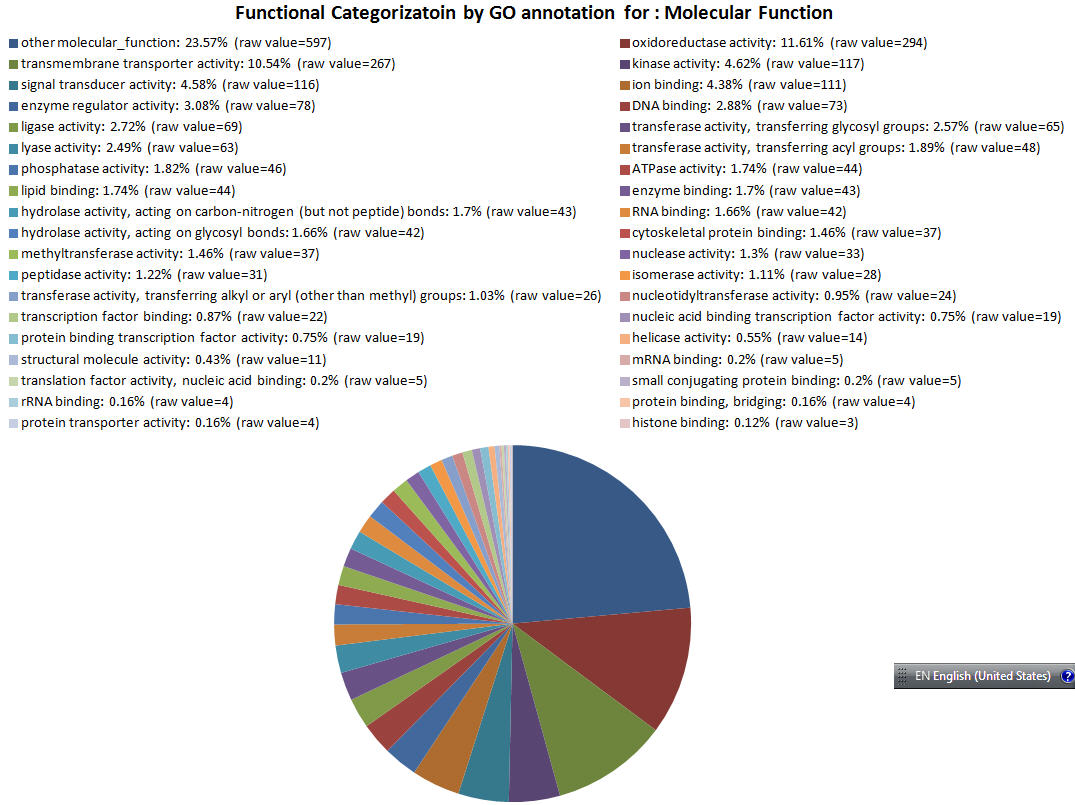
| GOID Link to GO |
Functional Category | GO Annotation Count to see GO terms |
|---|---|---|
| GO:0003674 | other molecular_function | 597 |
| GO:0016491 | oxidoreductase activity | 294 |
| GO:0022857 | transmembrane transporter activity | 267 |
| GO:0016301 | kinase activity | 117 |
| GO:0004871 | signal transducer activity | 116 |
| GO:0043167 | ion binding | 111 |
| GO:0030234 | enzyme regulator activity | 78 |
| GO:0003677 | DNA binding | 73 |
| GO:0016874 | ligase activity | 69 |
| GO:0016757 | transferase activity, transferring glycosyl groups | 65 |
| GO:0016829 | lyase activity | 63 |
| GO:0016746 | transferase activity, transferring acyl groups | 48 |
| GO:0016791 | phosphatase activity | 46 |
| GO:0016887 | ATPase activity | 44 |
| GO:0008289 | lipid binding | 44 |
| GO:0019899 | enzyme binding | 43 |
| GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 43 |
| GO:0003723 | RNA binding | 42 |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | 42 |
| GO:0008092 | cytoskeletal protein binding | 37 |
| GO:0008168 | methyltransferase activity | 37 |
| GO:0004518 | nuclease activity | 33 |
| GO:0008233 | peptidase activity | 31 |
| GO:0016853 | isomerase activity | 28 |
| GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups | 26 |
| GO:0016779 | nucleotidyltransferase activity | 24 |
| GO:0008134 | transcription factor binding | 22 |
| GO:0001071 | nucleic acid binding transcription factor activity | 19 |
| GO:0000988 | protein binding transcription factor activity | 19 |
| GO:0004386 | helicase activity | 14 |
| GO:0005198 | structural molecule activity | 11 |
| GO:0003729 | mRNA binding | 5 |
| GO:0008135 | translation factor activity, nucleic acid binding | 5 |
| GO:0032182 | small conjugating protein binding | 5 |
| GO:0019843 | rRNA binding | 4 |
| GO:0030674 | protein binding, bridging | 4 |
| GO:0008565 | protein transporter activity | 4 |
| GO:0042393 | histone binding | 3 |