T0392
PDZ domain of Rhophilin-2 Homo sapiens
Target sequence:
>T0392 RHPN2A, Homo sapiens, 109 residues
MHHHHHHSSGVDLGTENLYFQSMPRSIRFTAEEGDLGFTLRGNAPVQVHFLDPYCSASVAGAREGDYIVSIQLVDCKWLTLSEVMKLLKSFGEDEIEMKVVSLLDEVTF
Structure:
Determined by:
SGC
PDB ID: 2vsv
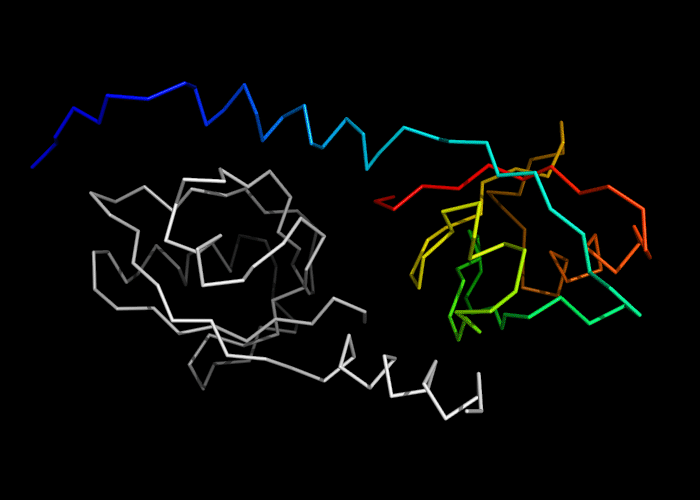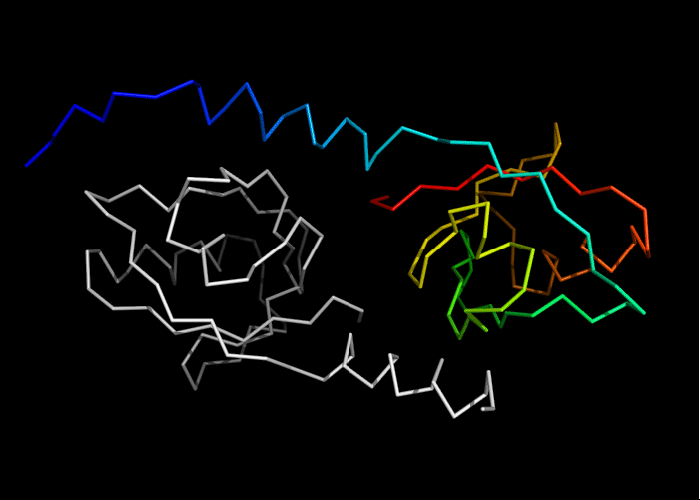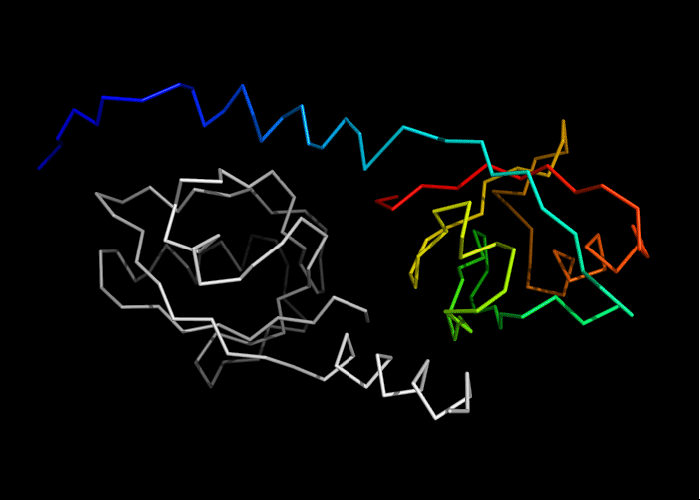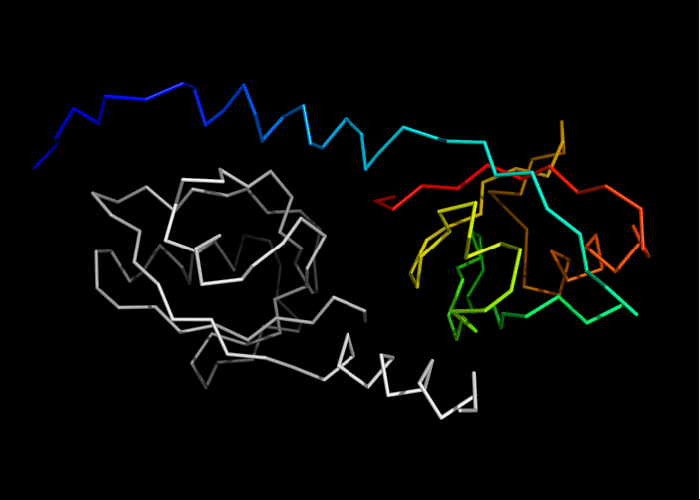
Ribbon diagram of 392: 2vsv chains A (white) and B (rainbow)
Domains: PyMOL of domains
Single domain protein. However, 1-23 (target numbers, and -23-0 in PDB numbers) are introduced in cloning, protrude away from the domain in 3D structure (interact with another chain, see above) and can be deleted for domain evaluation.
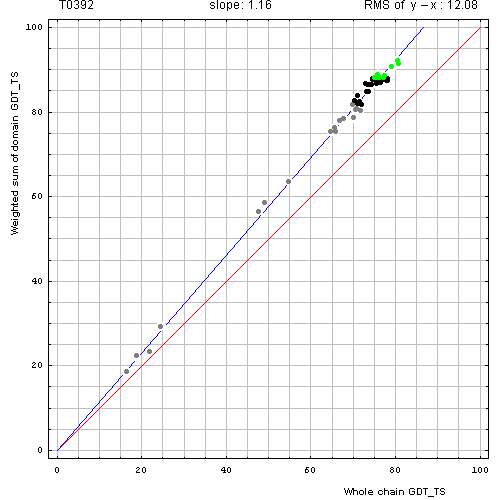
Correlation between GDT-TS scores for domain-based evlatuation (without N-terminal segment, y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
PDZ domain fold.
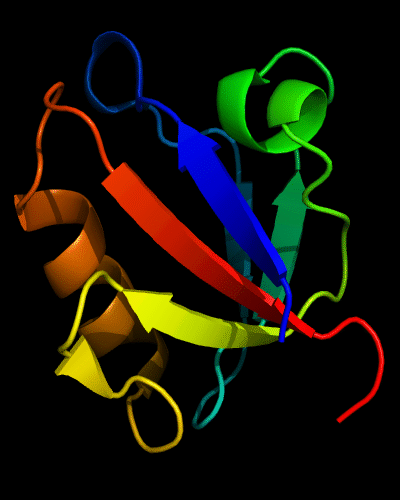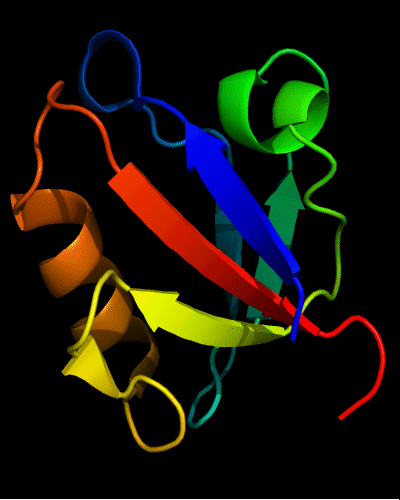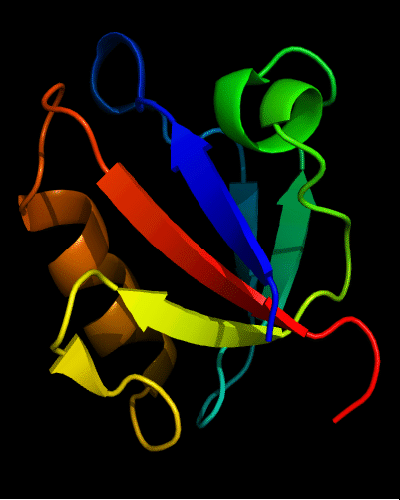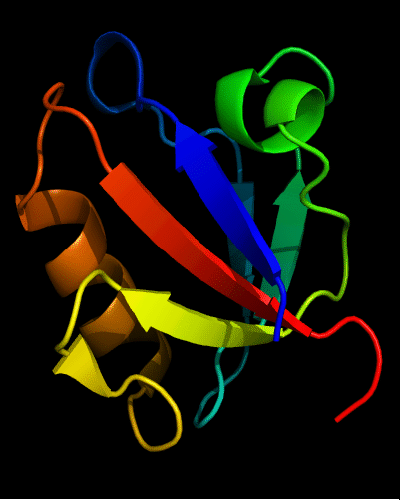
Cartoon diagram of 392: 2vsv chain B residues 514-595
CASP category:
Whole chain: Comparative modeling:medium.
1st domain(with N-terminal segment seq:1-23 removed): Comparative modeling:easy.
Closest templates:
Target sequence - PDB file inconsistencies:
The most complete chain B is missing only the C-terminus 105-DEVTF-109. PDB numbering is different from the target. PDB coordinates are from -22 to 0 and from 514 to 594, which corresponds to the target 1 to 104.
T0392 2vsv.pdb T0392.pdb PyMOL PyMOL of domains
T0392 1 MHHHHHHSSGVDLGTENLYFQSMPRSIRFTAEEGDLGFTLRGNAPVQVHFLDPYCSASVAGAREGDYIVSIQLVDCKWLTLSEVMKLLKSFGEDEIEMKVVSLLDEVTF 109 ~~~~~~~|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~ 2vsvA -15 -------SSGVDLGTENLYFQSMPRSIRFTAEEGDLGFTLRGNAPVQVHFLDPYCSASVAGAREGDYIVSIQLVDCKWLTLSEVMKLLKSFGEDEIEMKVVSLL----- 594
We suggest to evaluate this target as a single domain spanning whole chain. However, N-termial segment (target 1-23, pdb -15-0) is introduced in cloning, protrudes away from the domain in 3D structure (interact with another chain) and can be deleted for domain evaluation, resulting in this domain definition:
1st domain: target 24-104 ; pdb 514-594
Sequence classification:
PDZ signaling domain in CDD.
Server predictions:
T0392:pdb -15-594:seq 8-104:CM_medium; alignment
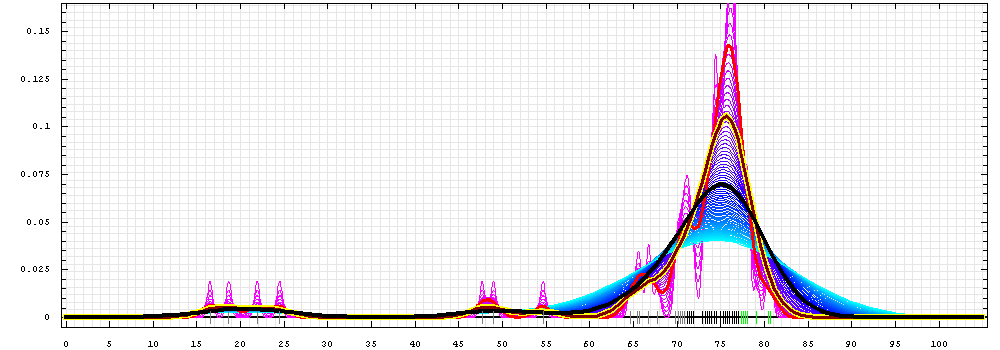
First models for T0392:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
392_1:pdb 514-594:seq 24-104:CM_easy; alignment
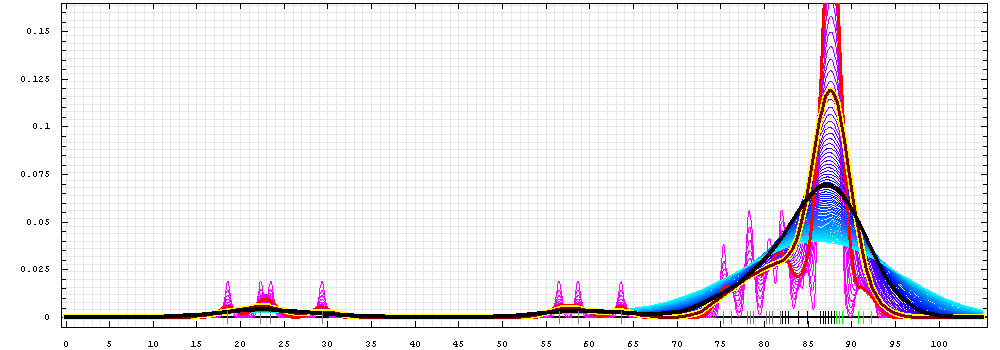
First models for T0392_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0392 | 392_1 | T0392 | 392_1 | T0392 | 392_1 | T0392 | 392_1 | ||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||
| 1 | pro−sp3−TASSER | 80.67 | 76.38 | 78.22 | 91.36 | 89.71 | 86.79 | 1.95 | 2.41 | 1.82 | 1.55 | 2.34 | 1.64 | 80.67 | 76.38 | 78.22 | 92.28 | 89.71 | 86.79 | 1.89 | 1.80 | 1.72 | 1.93 | 1.92 | 1.70 | pro−sp3−TASSER | 1 |
| 2 | MULTICOM−RANK | 80.41 | 79.72 | 76.57 | 92.28 | 91.46 | 85.86 | 1.87 | 3.28 | 1.42 | 1.81 | 2.68 | 1.41 | 80.41 | 79.72 | 76.57 | 92.28 | 91.46 | 85.86 | 1.79 | 2.70 | 1.25 | 1.93 | 2.31 | 1.39 | MULTICOM−RANK | 2 |
| 3 | Zhang−Server | 79.12 | 77.75 | 77.35 | 90.74 | 89.09 | 85.18 | 1.48 | 2.77 | 1.61 | 1.38 | 2.22 | 1.24 | 80.67 | 79.30 | 79.76 | 92.59 | 91.36 | 87.67 | 1.89 | 2.59 | 2.16 | 2.05 | 2.29 | 1.99 | Zhang−Server | 3 |
| 4 | MULTICOM−CMFR | 78.09 | 69.16 | 74.78 | 87.35 | 76.65 | 81.27 | 1.16 | 0.55 | 0.97 | 0.44 | 0.27 | 78.09 | 69.16 | 74.78 | 87.35 | 81.28 | 81.27 | 0.95 | 0.74 | 0.05 | 0.07 | MULTICOM−CMFR | 4 | |||
| 5 | FFASsuboptimal | 78.09 | 68.73 | 73.60 | 87.35 | 76.03 | 81.23 | 1.16 | 0.44 | 0.68 | 0.44 | 0.26 | 78.09 | 68.73 | 74.43 | 87.96 | 77.47 | 81.43 | 0.95 | 0.65 | 0.28 | FFASsuboptimal | 5 | ||||
| 6 | Phyre_de_novo | 78.09 | 68.47 | 75.76 | 87.96 | 76.85 | 82.73 | 1.16 | 0.37 | 1.22 | 0.61 | 0.64 | 78.09 | 71.56 | 75.76 | 87.96 | 82.51 | 82.73 | 0.95 | 0.49 | 1.02 | 0.28 | 0.34 | 0.37 | Phyre_de_novo | 6 | |
| 7 | FEIG | 77.83 | 66.84 | 72.96 | 87.35 | 75.82 | 81.67 | 1.08 | 0.53 | 0.44 | 0.37 | 77.83 | 72.42 | 72.96 | 87.65 | 79.32 | 81.67 | 0.86 | 0.72 | 0.23 | 0.16 | 0.02 | FEIG | 7 | |||
| 8 | BAKER−ROBETTA | 77.58 | 68.21 | 73.94 | 87.96 | 76.65 | 81.43 | 1.01 | 0.30 | 0.77 | 0.61 | 0.31 | 78.87 | 76.63 | 74.77 | 89.20 | 87.14 | 82.48 | 1.23 | 1.87 | 0.74 | 0.75 | 1.36 | 0.29 | BAKER−ROBETTA | 8 | |
| 9 | FALCON | 77.32 | 74.74 | 75.55 | 88.58 | 86.52 | 85.16 | 0.93 | 1.99 | 1.16 | 0.78 | 1.72 | 1.24 | 77.58 | 75.17 | 76.66 | 88.58 | 86.52 | 86.29 | 0.76 | 1.47 | 1.28 | 0.52 | 1.22 | 1.53 | FALCON | 9 |
| 10 | 3DShot2 | 77.06 | 73.97 | 71.84 | 88.27 | 84.57 | 81.10 | 0.85 | 1.79 | 0.25 | 0.69 | 1.34 | 0.23 | 77.06 | 73.97 | 71.84 | 88.27 | 84.57 | 81.10 | 0.58 | 1.14 | 0.40 | 0.79 | 3DShot2 | 10 | ||
| 11 | nFOLD3 | 77.06 | 68.13 | 73.21 | 87.65 | 76.95 | 81.48 | 0.85 | 0.28 | 0.59 | 0.52 | 0.33 | 77.58 | 75.00 | 73.30 | 88.89 | 86.42 | 82.53 | 0.76 | 1.42 | 0.32 | 0.64 | 1.20 | 0.30 | nFOLD3 | 11 | |
| 12 | huber−torda−server | 76.80 | 67.44 | 72.22 | 87.96 | 76.65 | 81.43 | 0.77 | 0.10 | 0.34 | 0.61 | 0.31 | 76.80 | 67.44 | 72.22 | 87.96 | 80.25 | 81.43 | 0.48 | 0.02 | 0.28 | huber−torda−server | 12 | ||||
| 13 | MUProt | 76.55 | 69.33 | 74.23 | 87.04 | 84.16 | 82.37 | 0.69 | 0.59 | 0.84 | 0.35 | 1.26 | 0.55 | 76.55 | 73.37 | 74.23 | 88.27 | 85.80 | 82.89 | 0.39 | 0.98 | 0.59 | 0.40 | 1.06 | 0.42 | MUProt | 13 |
| 14 | LEE−SERVER | 76.55 | 68.30 | 74.89 | 87.65 | 77.78 | 82.14 | 0.69 | 0.33 | 1.00 | 0.52 | 0.01 | 0.49 | 77.32 | 69.42 | 75.15 | 87.96 | 78.50 | 82.31 | 0.67 | 0.85 | 0.28 | 0.23 | LEE−SERVER | 14 | ||
| 15 | FAMSD | 76.55 | 67.53 | 72.42 | 87.65 | 76.75 | 81.10 | 0.69 | 0.13 | 0.39 | 0.52 | 0.23 | 76.55 | 69.24 | 72.42 | 87.65 | 81.38 | 81.10 | 0.39 | 0.07 | 0.16 | 0.09 | FAMSD | 15 | |||
| 16 | Phyre2 | 76.55 | 67.18 | 72.80 | 87.96 | 76.65 | 81.43 | 0.69 | 0.04 | 0.49 | 0.61 | 0.31 | 76.55 | 71.82 | 72.80 | 87.96 | 82.92 | 81.43 | 0.39 | 0.56 | 0.18 | 0.28 | 0.43 | Phyre2 | 16 | ||
| 17 | circle | 76.29 | 67.95 | 73.26 | 87.35 | 77.26 | 81.27 | 0.61 | 0.24 | 0.60 | 0.44 | 0.27 | 76.55 | 69.33 | 73.30 | 87.35 | 79.94 | 81.27 | 0.39 | 0.32 | 0.05 | circle | 17 | ||||
| 18 | 3Dpro | 76.29 | 67.35 | 73.45 | 87.65 | 76.95 | 81.32 | 0.61 | 0.08 | 0.65 | 0.52 | 0.29 | 76.29 | 67.35 | 73.45 | 87.65 | 76.95 | 81.32 | 0.30 | 0.37 | 0.16 | 3Dpro | 18 | ||||
| 19 | Poing | 76.29 | 66.92 | 72.84 | 87.96 | 76.65 | 81.43 | 0.61 | 0.50 | 0.61 | 0.31 | 76.29 | 71.65 | 72.84 | 87.96 | 82.92 | 81.43 | 0.30 | 0.52 | 0.19 | 0.28 | 0.43 | Poing | 19 | |||
| 20 | Phragment | 76.29 | 66.92 | 72.83 | 87.96 | 76.65 | 81.43 | 0.61 | 0.49 | 0.61 | 0.31 | 76.29 | 71.65 | 72.83 | 87.96 | 82.92 | 81.43 | 0.30 | 0.52 | 0.19 | 0.28 | 0.43 | Phragment | 20 | |||
| 21 | SAM−T08−server | 76.29 | 66.50 | 75.33 | 88.27 | 84.98 | 83.45 | 0.61 | 1.11 | 0.69 | 1.42 | 0.81 | 76.29 | 71.82 | 75.33 | 88.27 | 85.08 | 83.45 | 0.30 | 0.56 | 0.90 | 0.40 | 0.90 | 0.61 | SAM−T08−server | 21 | |
| 22 | HHpred2 | 76.03 | 68.99 | 73.19 | 87.35 | 78.91 | 81.29 | 0.53 | 0.50 | 0.58 | 0.44 | 0.24 | 0.28 | 76.03 | 68.99 | 73.19 | 87.35 | 78.91 | 81.29 | 0.20 | 0.29 | 0.05 | HHpred2 | 22 | |||
| 23 | mGenTHREADER | 76.03 | 66.67 | 71.63 | 87.96 | 76.65 | 81.43 | 0.53 | 0.20 | 0.61 | 0.31 | 76.03 | 66.67 | 71.63 | 87.96 | 76.65 | 81.43 | 0.20 | 0.28 | mGenTHREADER | 23 | ||||||
| 24 | MUSTER | 76.03 | 65.98 | 73.45 | 87.35 | 75.21 | 81.30 | 0.53 | 0.65 | 0.44 | 0.28 | 77.58 | 71.73 | 74.00 | 89.51 | 82.92 | 82.60 | 0.76 | 0.54 | 0.52 | 0.87 | 0.43 | 0.33 | MUSTER | 24 | ||
| 25 | PS2−server | 76.03 | 61.94 | 72.55 | 87.65 | 70.78 | 81.37 | 0.53 | 0.42 | 0.52 | 0.30 | 76.03 | 73.02 | 72.55 | 87.65 | 82.41 | 81.37 | 0.20 | 0.89 | 0.11 | 0.16 | 0.32 | PS2−server | 25 | |||
| 26 | Pcons_local | 75.77 | 68.90 | 70.45 | 88.89 | 84.36 | 82.86 | 0.45 | 0.48 | 0.87 | 1.30 | 0.67 | 76.03 | 71.56 | 70.45 | 88.89 | 84.98 | 82.86 | 0.20 | 0.49 | 0.64 | 0.88 | 0.41 | Pcons_local | 26 | ||
| 27 | YASARA | 75.77 | 68.90 | 76.37 | 88.27 | 80.04 | 84.97 | 0.45 | 0.48 | 1.37 | 0.69 | 0.46 | 1.19 | 75.77 | 68.90 | 76.37 | 88.27 | 80.04 | 84.97 | 0.11 | 1.20 | 0.40 | 1.10 | YASARA | 27 | ||
| 28 | keasar−server | 75.77 | 65.72 | 71.63 | 87.96 | 75.82 | 81.57 | 0.45 | 0.20 | 0.61 | 0.35 | 76.03 | 67.44 | 72.27 | 87.96 | 77.88 | 82.07 | 0.20 | 0.03 | 0.28 | 0.15 | keasar−server | 28 | ||||
| 29 | forecast | 75.52 | 66.15 | 72.69 | 87.04 | 75.72 | 81.56 | 0.38 | 0.46 | 0.35 | 0.35 | 75.52 | 66.15 | 72.69 | 87.04 | 83.54 | 81.58 | 0.02 | 0.15 | 0.57 | forecast | 29 | |||||
| 30 | PSI | 75.52 | 65.89 | 73.07 | 86.73 | 75.21 | 81.09 | 0.38 | 0.55 | 0.27 | 0.23 | 76.29 | 72.34 | 73.07 | 88.89 | 84.36 | 81.85 | 0.30 | 0.70 | 0.26 | 0.64 | 0.75 | 0.08 | PSI | 30 | ||
| 31 | MULTICOM−REFINE | 75.26 | 71.48 | 73.09 | 88.27 | 83.95 | 83.32 | 0.30 | 1.15 | 0.56 | 0.69 | 1.22 | 0.78 | 76.55 | 72.25 | 74.08 | 88.27 | 83.95 | 83.32 | 0.39 | 0.68 | 0.55 | 0.40 | 0.66 | 0.56 | MULTICOM−REFINE | 31 |
| 32 | Fiser−M4T | 75.26 | 68.39 | 69.18 | 87.65 | 79.42 | 81.20 | 0.30 | 0.35 | 0.52 | 0.33 | 0.26 | 75.26 | 68.39 | 69.18 | 87.65 | 79.42 | 81.20 | 0.16 | Fiser−M4T | 32 | ||||||
| 33 | ACOMPMOD | 75.00 | 68.64 | 73.64 | 88.27 | 82.92 | 86.22 | 0.22 | 0.41 | 0.69 | 0.69 | 1.02 | 1.50 | 75.00 | 68.64 | 73.64 | 88.27 | 82.92 | 86.22 | 0.42 | 0.40 | 0.43 | 1.51 | ACOMPMOD | 33 | ||
| 34 | OLGAFS | 75.00 | 66.41 | 70.86 | 87.96 | 76.44 | 81.43 | 0.22 | 0.01 | 0.61 | 0.31 | 75.00 | 66.41 | 70.86 | 87.96 | 76.44 | 81.43 | 0.28 | OLGAFS | 34 | |||||||
| 35 | Pcons_dot_net | 74.48 | 71.91 | 73.59 | 87.65 | 85.19 | 86.06 | 0.06 | 1.26 | 0.68 | 0.52 | 1.46 | 1.46 | 75.77 | 71.91 | 73.59 | 88.89 | 85.19 | 86.06 | 0.11 | 0.59 | 0.41 | 0.64 | 0.93 | 1.46 | Pcons_dot_net | 35 |
| 36 | Pcons_multi | 74.48 | 71.91 | 73.59 | 87.65 | 85.19 | 86.06 | 0.06 | 1.26 | 0.68 | 0.52 | 1.46 | 1.46 | 79.90 | 74.91 | 74.68 | 89.81 | 86.52 | 86.06 | 1.61 | 1.40 | 0.72 | 0.99 | 1.22 | 1.46 | Pcons_multi | 36 |
| 37 | FUGUE_KM | 74.48 | 71.73 | 72.27 | 87.96 | 85.70 | 84.96 | 0.06 | 1.21 | 0.36 | 0.61 | 1.56 | 1.19 | 74.48 | 71.73 | 72.27 | 87.96 | 85.70 | 84.96 | 0.54 | 0.03 | 0.28 | 1.04 | 1.10 | FUGUE_KM | 37 | |
| 38 | FFASstandard | 74.48 | 65.21 | 68.29 | 87.96 | 76.85 | 81.31 | 0.06 | 0.61 | 0.28 | 74.48 | 65.21 | 71.44 | 87.96 | 76.85 | 81.43 | 0.28 | FFASstandard | 38 | ||||||||
| 39 | FFASflextemplate | 74.48 | 65.21 | 68.29 | 87.96 | 76.85 | 81.31 | 0.06 | 0.61 | 0.28 | 74.48 | 65.21 | 68.36 | 87.96 | 76.85 | 81.43 | 0.28 | FFASflextemplate | 39 | ||||||||
| 40 | GeneSilicoMetaServer | 74.23 | 66.15 | 67.04 | 86.42 | 84.36 | 79.20 | 0.18 | 1.30 | 74.48 | 71.73 | 72.32 | 87.65 | 85.19 | 84.41 | 0.54 | 0.04 | 0.16 | 0.93 | 0.92 | GeneSilicoMetaServer | 40 | |||||
| 41 | CpHModels | 74.23 | 60.48 | 70.78 | 86.42 | 69.96 | 83.03 | 0.18 | 0.71 | 74.23 | 60.48 | 70.78 | 86.42 | 69.96 | 83.03 | 0.47 | CpHModels | 41 | |||||||||
| 42 | MUFOLD−Server | 73.97 | 69.33 | 73.33 | 86.42 | 81.28 | 80.74 | 0.59 | 0.62 | 0.18 | 0.70 | 0.14 | 74.48 | 69.59 | 73.38 | 87.35 | 81.28 | 81.39 | 0.35 | 0.05 | 0.07 | MUFOLD−Server | 42 | ||||
| 43 | RAPTOR | 73.71 | 71.65 | 71.05 | 84.88 | 82.41 | 80.07 | 1.19 | 0.05 | 0.92 | 78.09 | 73.28 | 76.65 | 88.89 | 84.77 | 85.48 | 0.95 | 0.96 | 1.28 | 0.64 | 0.84 | 1.27 | RAPTOR | 43 | |||
| 44 | MULTICOM−CLUSTER | 73.45 | 69.50 | 71.78 | 86.42 | 81.69 | 81.57 | 0.64 | 0.23 | 0.18 | 0.78 | 0.35 | 77.32 | 73.11 | 75.49 | 87.96 | 83.75 | 82.60 | 0.67 | 0.91 | 0.95 | 0.28 | 0.61 | 0.33 | MULTICOM−CLUSTER | 44 | |
| 45 | COMA | 73.45 | 68.47 | 68.08 | 86.42 | 80.66 | 80.01 | 0.37 | 0.18 | 0.58 | 73.45 | 68.47 | 68.08 | 86.42 | 80.66 | 80.01 | COMA | 45 | |||||||||
| 46 | rehtnap | 73.20 | 69.42 | 68.57 | 84.88 | 80.35 | 78.88 | 0.61 | 0.52 | 74.48 | 71.05 | 68.92 | 86.42 | 81.89 | 79.53 | 0.35 | 0.20 | rehtnap | 46 | ||||||||
| 47 | BioSerf | 72.94 | 60.74 | 70.64 | 86.73 | 72.33 | 79.41 | 0.27 | 72.94 | 60.74 | 70.64 | 86.73 | 72.33 | 79.41 | BioSerf | 47 | |||||||||||
| 48 | fais−server | 71.91 | 67.27 | 67.55 | 81.79 | 76.44 | 77.64 | 0.06 | 76.03 | 67.27 | 72.07 | 87.65 | 77.26 | 81.41 | 0.20 | 0.16 | fais−server | 48 | |||||||||
| 49 | GS−KudlatyPred | 71.65 | 65.12 | 63.56 | 80.25 | 73.05 | 71.21 | 71.65 | 65.12 | 63.56 | 80.25 | 73.05 | 71.21 | GS−KudlatyPred | 49 | ||||||||||||
| 50 | pipe_int | 71.39 | 63.32 | 73.88 | 82.41 | 77.88 | 80.49 | 0.75 | 0.03 | 0.08 | 77.58 | 72.25 | 78.00 | 87.96 | 84.88 | 84.53 | 0.76 | 0.68 | 1.66 | 0.28 | 0.86 | 0.96 | pipe_int | 50 | |||
| 51 | Frankenstein | 71.13 | 67.70 | 68.44 | 82.10 | 78.19 | 77.93 | 0.17 | 0.09 | 75.26 | 68.56 | 75.18 | 87.04 | 80.35 | 83.30 | 0.86 | 0.56 | Frankenstein | 51 | ||||||||
| 52 | SAM−T02−server | 71.13 | 61.86 | 60.20 | 83.95 | 73.25 | 72.91 | 71.13 | 67.27 | 61.12 | 83.95 | 77.16 | 72.91 | SAM−T02−server | 52 | ||||||||||||
| 53 | mariner1 | 70.88 | 62.11 | 63.85 | 80.86 | 69.14 | 70.99 | 76.29 | 70.79 | 73.87 | 88.27 | 82.72 | 82.23 | 0.30 | 0.28 | 0.49 | 0.40 | 0.38 | 0.21 | mariner1 | 53 | ||||||
| 54 | METATASSER | 70.62 | 63.57 | 69.99 | 80.56 | 73.35 | 78.53 | 77.06 | 71.05 | 75.31 | 89.51 | 82.92 | 84.38 | 0.58 | 0.35 | 0.90 | 0.87 | 0.43 | 0.91 | METATASSER | 54 | ||||||
| 55 | Distill | 70.36 | 57.82 | 61.22 | 82.72 | 67.69 | 72.04 | 70.62 | 63.92 | 64.15 | 83.03 | 70.68 | 75.02 | Distill | 55 | ||||||||||||
| 56 | GS−MetaServer2 | 70.10 | 63.23 | 61.29 | 78.70 | 71.09 | 70.41 | 73.97 | 67.70 | 68.73 | 87.65 | 81.07 | 80.71 | 0.16 | 0.02 | GS−MetaServer2 | 56 | ||||||||||
| 57 | panther_server | 69.84 | 59.88 | 62.10 | 81.79 | 71.09 | 72.86 | 72.42 | 69.16 | 66.95 | 84.57 | 80.45 | 77.82 | panther_server | 57 | ||||||||||||
| 58 | SAM−T06−server | 67.78 | 63.49 | 64.61 | 78.39 | 74.07 | 68.73 | 70.10 | 67.27 | 64.61 | 83.64 | 79.53 | 76.34 | SAM−T06−server | 58 | ||||||||||||
| 59 | HHpred4 | 66.75 | 63.66 | 63.07 | 78.09 | 71.30 | 73.19 | 66.75 | 63.66 | 63.07 | 78.09 | 71.30 | 73.19 | HHpred4 | 59 | ||||||||||||
| 60 | HHpred5 | 66.75 | 63.66 | 63.07 | 78.09 | 71.30 | 73.19 | 66.75 | 63.66 | 63.07 | 78.09 | 71.30 | 73.19 | HHpred5 | 60 | ||||||||||||
| 61 | 3D−JIGSAW_V3 | 65.72 | 60.83 | 65.07 | 75.31 | 69.14 | 71.60 | 70.10 | 67.01 | 71.92 | 80.86 | 77.78 | 80.10 | 3D−JIGSAW_V3 | 61 | ||||||||||||
| 62 | COMA−M | 65.46 | 59.97 | 64.34 | 76.23 | 69.24 | 73.85 | 72.68 | 68.73 | 67.51 | 84.26 | 80.56 | 77.55 | COMA−M | 62 | ||||||||||||
| 63 | 3D−JIGSAW_AEP | 64.69 | 60.22 | 69.59 | 75.31 | 69.96 | 76.29 | 65.98 | 60.48 | 69.59 | 75.93 | 69.96 | 76.57 | 3D−JIGSAW_AEP | 63 | ||||||||||||
| 64 | FALCON_CONSENSUS | 54.64 | 43.81 | 50.10 | 63.58 | 51.65 | 57.18 | 54.64 | 43.81 | 50.10 | 63.58 | 51.65 | 57.18 | FALCON_CONSENSUS | 64 | ||||||||||||
| 65 | MUFOLD−MD | 48.97 | 42.95 | 47.22 | 58.64 | 51.44 | 55.64 | 48.97 | 42.95 | 47.22 | 58.64 | 51.44 | 55.64 | MUFOLD−MD | 65 | ||||||||||||
| 66 | FOLDpro | 47.68 | 40.29 | 40.02 | 56.48 | 47.63 | 48.47 | 64.95 | 59.19 | 57.38 | 73.77 | 67.80 | 63.89 | FOLDpro | 66 | ||||||||||||
| 67 | RBO−Proteus | 24.48 | 21.65 | 32.68 | 29.32 | 25.93 | 36.72 | 24.48 | 21.65 | 32.68 | 29.32 | 25.93 | 37.08 | RBO−Proteus | 67 | ||||||||||||
| 68 | mahmood−torda−server | 21.91 | 15.38 | 28.43 | 23.46 | 17.49 | 28.98 | 21.91 | 17.27 | 28.43 | 23.46 | 18.42 | 28.98 | mahmood−torda−server | 68 | ||||||||||||
| 69 | RANDOM | 18.64 | 15.40 | 18.64 | 22.31 | 18.03 | 22.31 | 18.64 | 15.40 | 18.64 | 22.31 | 18.03 | 22.31 | RANDOM | 69 | ||||||||||||
| 70 | schenk−torda−server | 16.50 | 14.09 | 22.13 | 18.52 | 15.54 | 25.13 | 18.81 | 15.98 | 22.13 | 21.30 | 19.14 | 25.13 | schenk−torda−server | 70 | ||||||||||||
| 71 | BHAGEERATH | BHAGEERATH | 71 | ||||||||||||||||||||||||
| 72 | LOOPP_Server | LOOPP_Server | 72 | ||||||||||||||||||||||||
| 73 | Pushchino | Pushchino | 73 | ||||||||||||||||||||||||
| 74 | test_http_server_01 | test_http_server_01 | 74 | ||||||||||||||||||||||||