T0489
metal-dependent hydrolase from Klebsiella pneumoniae
Target sequence:
>T0489 Putative Metal-dependent Hydrolase, Klebsiella pneumoniae, 266 residues
MMFKWPWKADDESGNAEMPWEQALAIPVLAHLSSTEQHKLTQMAARFLQQKRLVALQGLELTPLHQARIAMLFCLPVLELGIEWLDGFHEVLIYPAPFIVDDEWEDDIGLVHNQRVVQSGQSWQQGPVVLNWLDIQDSFDASGFNLVVHEVAHKLDTRNGDRASGVPLIPLREVAGWEHDLHAAMNNIQDEIDLVGESAASIDAYAATDPAECFAVLSEYFFSAPELFAPRFPALWQRFCHFYRQDPLARRRENGLQDEGDRRIVH
Structure:
Determined by:
JCSG
PDB ID: 3dl1
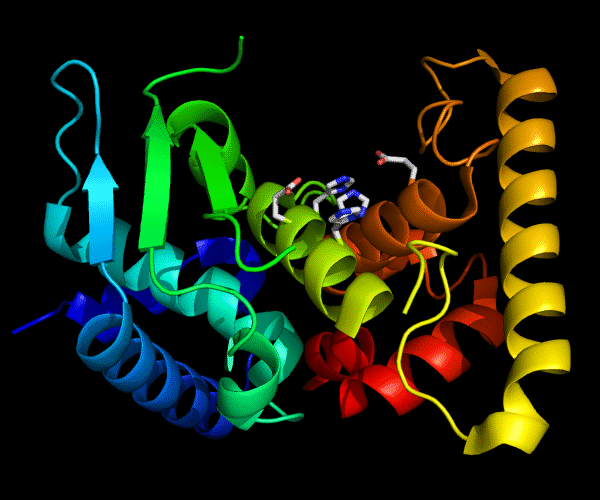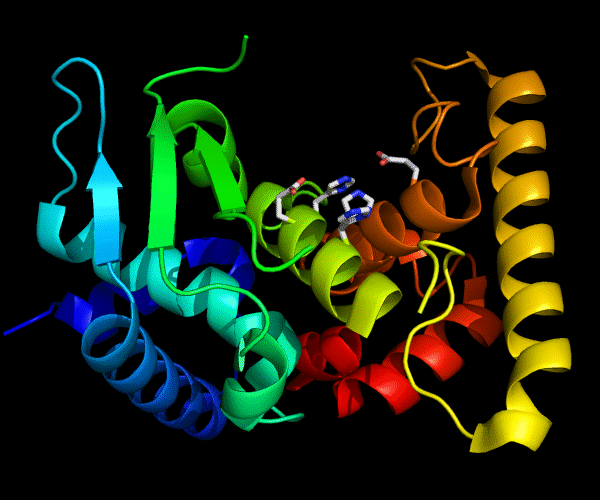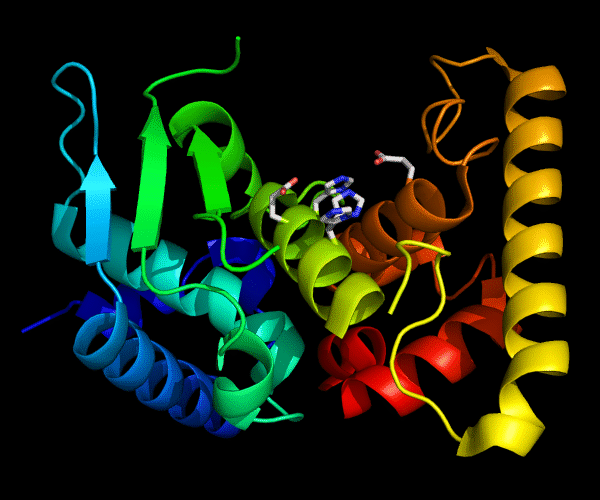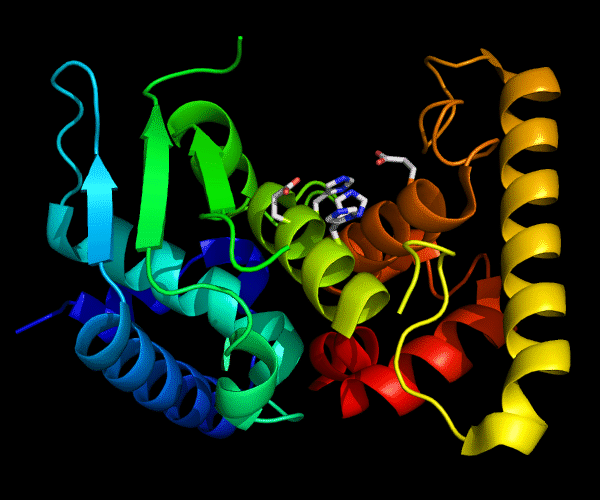
Cartoon diagram of 489: 3dl1. Active site residues shown as "sticks".
Domains: PyMOL of domains
Single domain protein for evaluation, but evolutionarily two domains. β/α and a C-terminal helical array. Poorly ordered structure with the most interesting edge β-strand from the catalytic site being disordered.
Structure classification:
Zincin fold.
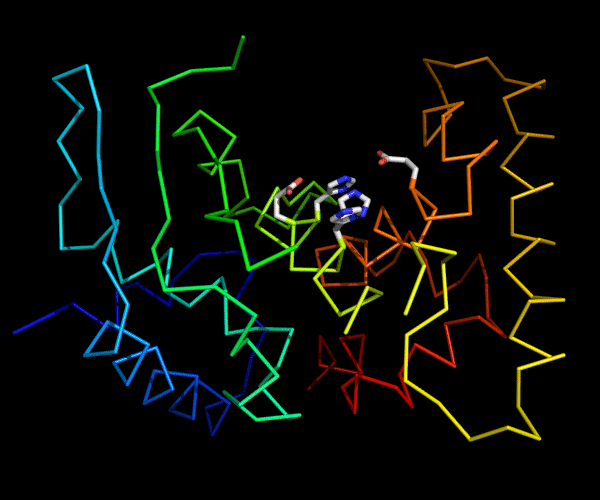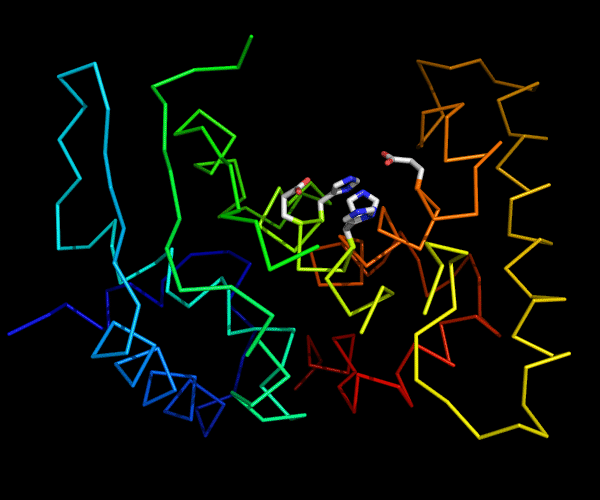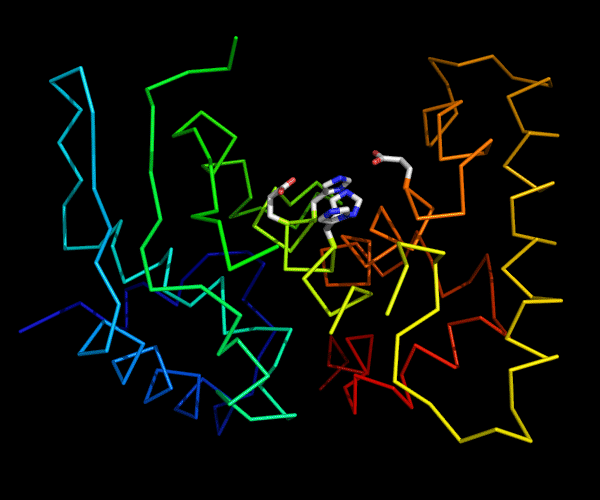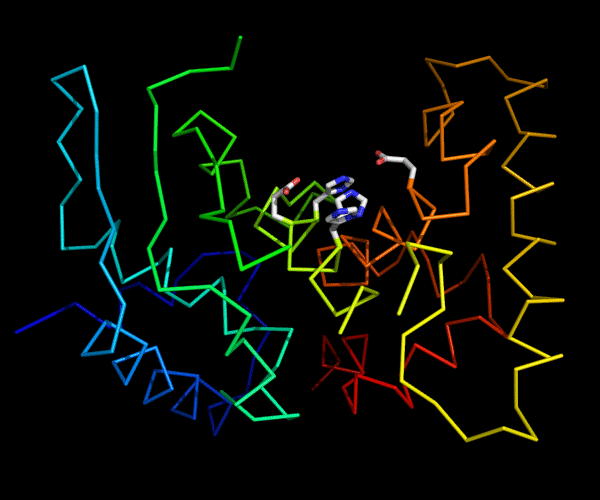
Ribbon diagram of 489: 3dl1. Active site residues shown as "sticks".
CASP category:
Fold recognition.
Closest templates:
1j7n.
Conservation: 6 6 7 5 65 5 6 5 6 556 7 5 5 555 655 88 58 65 5 5 5 86 97798 9 67 7 5 5 55 6 5 gi_110637843_ref_YP_67805 1 --------------------------MIKLF----------------IVAC----GYIIYKLG----RYCKEN-EYYMAS----LLMLQYH---KSLSAERQQILY-TNFTYYKKLSEKKKLEFEKRVQHFLTNKSFIASDGFE-ITEQMKVMIAATAVQILFCRK-----AYYLSSFDFIELFEEVPV---------------------HVEVMRKK-RIIQISWNEF-KKGF-------DSLIDG--YNPGLKIMAMALDLEHK---FGEKS-LFSQ--------QTYKAFQKIYKQEAEKYILS-----------------------GKSKYDHYNKVDRTEYFAVAVEYFFERPEHFNVNQPAMYLALSKLLRQDPLG-----MYTYKEKVFE------------
gi_110639693_ref_YP_67990 1 --------------------------MAPDQKTS--------PWWFIGTVIVIVSAIIINQCI-----------IYEIHL----SKIFRYL---LPLSSSRCAILE-ERFYYYNRLSKDDQRAFRKRVHHFLVNKKFTSEDSIE-ITEEMKVMIAAAATLIMFHLE-----AYYLSHFDAIHISNNELE---------------------SLQHMHKA-KEVEIPWDDF-RAGF-------DSFHDG--YNPGLKILAMAFTLEHQL-NNLSSK-MFRF--------NRYYDLMQLYKNEAEKYIAS-----------------------GKSKYKEYSEVDRNEYFAVAVEYFFERPGHFYANQPAMYIALTKVLRQDPLG-----MYTYKRK---------------
gi_89891275_ref_ZP_012027 1 --------------------------M-----------------------------------------------PIFNLF------RKKTP---APSTDHWDDILL-KYVSFFEKLEARDRLFFKLRMQAFLDEIEIETI-ETE-PTETDALLIAASAIIPVFQFP-----DWHYSNLKTVILLPEHFNTDL---EFEGHGNGRRIF--GMVGTGKWN-HKMILSKEAL-YHGF-------KNDTDK--LNTAIHEFVHLIDKMDGT-IDGIPH-VLLEQ-------SYVLPWINLMHEEMERINK------------------------DKSDIRQYGGSSQIEFFAVAAEYFFSRPKLMKRKHPDIYQMLSKCFTPDEE----------------------------
gi_86131502_ref_ZP_010501 1 --------------------------MIILF----------------LLA-----GLALFVFM-----------GYGLFT----LSRKRTP---DPFPPSWESLLS-DEVLFYKELTPKKKEFFKKRMMAFLTEVNVEAV-GFD-LSDKDKVFVAASAVIPIFNFG-----DWHYNNLDTVLIYPDYFNANL---EYHSEGEDRIIG--GMVGTGQFE-NHMILSRKAL-HFGF-------NNKTDK--GNTAIHEFVHLLDKMDGE-VDGIPEQLLGK--------ENIVPWLDMMHTEMEAINN------------------------DTSDIRKYGGTSKIEFFAVAAEYFFERPDLFKRKHPELYKMLEVCFVTGY-----------------------------
gi_149277260_ref_ZP_01883 1 --------------------------MMESF--------------FILI--------PIFLLV-----------LYFIFR----KDNSHNY---VNLSEFDRKLLF-DHVHYYQELTDEGKLRFEQKVVAFLSDVRLEGV-GTE-ITVLDKLLVASSAIIPVFGFP-----DWKYRNLSTVVLYPDTFNKDF---QFEGGDRSIL----GMVGTGFMN-GQMVLSRAAL-QQGF-------SNKADK--ENTAIHEFVHLLDKSDGA-TDGVPE-NLMKH-------EYTLPWLQMIHAEMQRIEK------------------------GKSDINPYAITNEAEFFAVVSEYFFEKPEQFKEKHPQLYSGLSSIFMQDPAE-----KNL-------------------
gi_126648834_ref_ZP_01721 1 --------------------------MKIFT----------------LLF-----AIFLVV---------LLV-WTIYKS--KVKKSGLKV--PELFPEKWRRFLE-SEVQFYSRLSPEEKSTFEADIQRFLKRIRITGV-KCE-VTDEDRLLVASSAVIPIFRFP-----EWEYKNLCEVLLYPDLFTEKF---DFTGD----HRYTSGMVGSGGPMNNVVIFSKPAL-RQGF-------ENRSDK--FNVGIHEFVHLFDKQDGS-IDGIPA-IIMKN-------QSVLPWLHLIKSNTSEILS------------------------GKSDINPYGATNNQEFLAVVSEYFFERPKLFQKKHPELYRILSQVFQNDLAK-----RN--------------------
gi_110636952_ref_YP_67715 1 --------------------------MFPII----------------VIA-----VLLFGL---------LFLFTQKKNK----KVKIVLA---QPFPEAWHAYLL-QEVKFYAELSADDQVRFQNKVKKFFSDTAITGVKDLE-VTDELKLLVAASAIIPVFLFD-----GWEYVNLSEVIIYDGAVEPNQ---TNDAESDGTLL---GQVRPLQTR-HVMLLSKQFL-VQGF-------ESMNGK--SNVGIHEFAHLIDQADGT-FDGIPK-AFMPE-------ELLKPWTKVMYAEIEKIAE------------------------NRSDINPYALTNHAEFFAVVCEYFFENPEKFQEKHPQLYDLMNQIFKREEAL-----TADGID----------------
gi_126663241_ref_ZP_01734 1 --------------------------MNPPPVEHELFLADYIAMFVVVVPFAAIFFVFAFYVIE---IIYLRN-YKKPLL----VFANIQF---KNLTESQKHILA-ANFNFYNNLKPKYKRYFEHRVSMFISNYEFLGR-DIS-VTEEMKITISATYVMLTFGMR-----DYLNPLFKTILIFPDIYYSSQ---TDNYH-----------KGEFNPRLKNIVFSWKHF-KEGI-------DITNDN--LNLGLHEFTHAFHIQSLK-SEKATF-VLFN--------ESLQSLFKLVSNPQMKQKLI-----------------------ESGFLRDYAYENQFEFVAVLLEYFFESPQEFKTKFPSIFLRVKHMINFNEKY-----FITSEF----------------
gi_146299580_ref_YP_00119 1 --------------------------MLFLF---------------LGGILF--GLIIVFRIIE---PTYLLL-FNKPLF----IFWHPIL---NKMNGADKTILR-REFPFYLNLSDKKKGYFEHRVKCFIDNYNFDGK-DIE-VTQEMKLIIAGTYVMLTFGMR-----TYLIHLFENIVIYPSVYYSTI---NDAYH-----------KGEYNPRMKTIVFSWEDF-LSGH--------QTKDN--INLGLHEFTHVLHFHSRK-RADPNA-IIFY--------DEFTEIEKYFDKEDLTNRLK-----------------------EKQYFRDYAYTNKFEFLAVILEHFFETPEIFKKEFPELFENVKTMINFKEEV---------------------------
gi_83857620_ref_ZP_009511 1 --------------------------MVFGK---------------FLAIAI--ILIIIYYLVKSAETLYVIK-FKKPIY----THFYLFK---KKLPESSITILE-NQFEFYGLLNTSQKTFFQHRLATFLADKEFFGREDLE-VTEEMKVLIGATAIMLTFGMR-----NYYLGILDRVIIYPNTYYSNI---TAAYH-----------KGEFNPRLKALVISWKDF-KEGY-------ADTTDN--LNLGIHEFTHILQLNSKK-FRDVSA-SIFS--------DGYTEIEQILKDDLYRNNLM-----------------------ASDYFRNYAYTNKYEFLAVLIENFIETPKDFKMNFPQLYSKVRQMLNYNFAG-----Y---------------------
gi_126646744_ref_ZP_01719 1 --------------------------MLLYYLF-------------------------------------SGR-FWLFEE----LRLSIMR---RRLKQSDLDILE-SRFPYFARLSNKHKREFCRKLEEILTIKTFIGRGGVQAVSQEMEVLIGATIVMVTFGWQ-----RVRLPHFKKILIYPNAYYSTI---SKTYH-----------KGEVNPRHGIIVVSWEAF-LEGL-------ENQKDG--LNLGIHEIAHALKLENQI-RNNSES-DFFNP-------QAYRDFHSFCASEIRKISSD-----------------------IPGFFRKEGALNEHEFFAVALENFFERPHQFFQYNSKLYGTLVQLLRQDPRV-----WIKEIA----------------
gi_110638647_ref_YP_67885 1 --------------------------M--------------------------------------------------------------------ISTEEIYTVLL-FEFPYFKKINEEKRQQLCLRTKRFIEETNFIPRKGIE-LTNRMVILISACSQQLTLGFS----NHYNYTYFEKIIVYPEKYLSTV---TEKYH-----------TGEMNTA-GIVVLSWEDF-YKGI-------KIDNDA--HNVGLHEFAHALEFMDIA-NKDV------------------NEVFSACLDKFTVLADQYLQHQ-----------------PDKPLFRSYATTNLSEFFAVATEYYFEAPYEFSQQEPELFDVLHKAYQQNTVP-----KASKPKLLAFPKPEEKDLLFGH
gi_168703525_ref_ZP_02735 1 --------------------------------------------MIF---------------------------SWLRAR----RRRKLLA---NAFPVRYAAIIE-RNVGHEALLPEPLRARLRDATTILLSEKEWLGRGGLF-VSEEMRVTIAAQAALLLLGRE-----HEYFDSVREVVVFPTTFQTPR----PEDGWEDDFLSDTISSGQAVYT-GTVLLAWDEV-QREG-------RDPAGG--YNVVLHEFAHQLDFAEGL-AGRAPR-LGDR--------ELEARWKYVMSVAFADHRRAVKAN------------------EPELFFTPHAADDELEFFADLTEAFFCRPYDLRNLYPELYRLLGAYYRLDPAA-----WAWPV-----------------
gi_158340234_ref_YP_00152 1 --------------------------------------------------------------------------------------MTIHK---KIFISDWEKILE-NNIPIYNFLPSSLQSKLKDLILIFLDDKRFFGCDGLE-ITDEIRITISAQACLLIINRK-----GKTYPLLSSILIYPDSINKKV---ERIENQFVVTDMRRSWLGDSNARKGQIRISWKHA-QLGA-------LNWKDG--QNLVLHEFAHQLDQEDSC-ADGVPI-LNCLNP-----QHAYMTWNKIITDEYKRLQSAVENN-------------------MVTVIDEYGATDLAEFFAVATEVFFEKPIPMKLNHSKLYKILSDFYGLDPAE-----WSYFSQ----------------
gi_157961121_ref_YP_00150 1 --------------------------ML------------------AIFLIL--LFSISAIA------WIASA-SWRARR----RRKLISQ---QPFPHEWREILK-RRIPFFRTLPADLQLQLKKHIQVFIAEKRFVGCDDLK-ITDDIRVTVAAQACLLLLNRT-----TDFYPRLKEILIYPSVFIVEN---QQHHGAGVVSEQRRVLSGESWQH-GRVILSWQTT-KHDA-------AVPDDG--SNVVIHEFAHQLDQEDGS-ANGAPI-LQHS--------KDYASWSEVLMQEFQALVRASQYG-------------------EHSLFSYYGATNPAEFFAVVSEVFFEQPHDFIALHPALYRELSAFYQLDPVN-----WH--------------------
gi_157374557_ref_YP_00147 1 --------------------------ML------------------GIILVS--IIGIAAIS------WIASH-SWRVER----QRKQIIK---HAFPKRWRDILK-RRMPYFRTLPTDLQLQLKQLIQVFINEKQFVGCDGIE-IDDEIRVTIAAQACLLLLNRH-----TDFYPNLKQILVYPSIFIVNN---KEQRDGGVISERQRILSGESWQQ-GKVILSWQTT-QADA-------ASPGDG--SNVVLHEFAHQLDQEAGD-ANGAPL-LSRV--------SDYSTWSEVLSEEYETLRLDAQQN-------------------TPSIFSYYGATSPAEFFAVITEVFFERPEEFYLQHKSLYKELSVFFNLDPIN-----WQ--------------------gi_149113867_ref_ZP_01840 1 --------------------------ML------------------AIVITL--ILGVGAIA------WLLSS-RWRKTL----HRRRVTA---KPFPKEWRSILK-KRLPYFHALPTDLQLQLKRLIQIFIDEKEFVGCDDFE-ITDEVRVTIAAQACLLLLNRK-----TDFYPKLKQILVYPDAFIVNN---QHRDTSGIVWEQRNVLAGESWEY-GKVVLSWKNT-LDGA-------ADPHDG--NNVVIHEFAHQLDQEDGQ-ANGAPI-LQHF--------ADYSTWSEVLSKAFTQLQHSANFG-------------------TPSLFNYYGATNPAEFFAVITEVFFEKPEDFYQQHPDLYRELSRFYQLDPIN-----WH--------------------gi_89902223_ref_YP_524694 1 --------------------------MP------------------ALIIS---ALALLFVL------WLLGE-PYLTER----KRQRIRA---QPFPAAWREILK-RRVPYVRALPADLQLQLKRHIQVFVAEKTFIGCAGLE-ITDEMRVTIAAQACLLILNRP-----GDYYPNLRQILVYPGSFVVHQ---AHTDDSGVAHQGRQILSGESWSE-GQVILSWQDT-LAGA-------ATPTDG--QNVAIHEFAHQLDQETGP-ANGAPL-LARR--------AHYARWSQVLGAEFDALRTRASPD-------------------EASLFSDYGATDPAEFFAVISEVFFEQPQRMAEEHPALYHELAQFYRLDPLS-----W---------------------
gi_83646881_ref_YP_435316 1 -----MSWKELAPFIVFLILAASAVVMV----------------------------------------------MWYPNQ----RLRKIRS---APFPETWEVILE-ANLPVYKRLPDNLKHQLQGLINVFLHQKTFYGCAGLE-ITDEMRVTIAGEACLLLLNRS-----TNEYNDLRFILVYPEAFRAER---EVINDDGTAAVSSHGLLGESWNN-GKVILSWDDV-IAGA-------RDFTDG--HNVVLHEFAHQLDGESGS-TNGAPL-LRTP--------TSYRSWARVLSDEFNILRDSAFKG-------------------KKSVMDNYGATNPAEFFAVATETFYEKPKEMAEKHPELFRELRNYYCVDPRD-----WV--------------------
gi_162451606_ref_YP_00161 1 --------------------------ML----------------------------------------------AFFRKR----RRDAVRR---ESFPPAWREILD-RNVPCFGRLPAEDQQELLGHIQVFLDEKPFEGCGGLE-ITDEIRVTVAAQACLLLLHRE-----TDYYPELEVILVYPSTYVARG---REVREGGVVVEGDHARLGESWSR-GSVVLAWDGV-LAGA-------ADMSDG--HNLVLHEFAHQLDQEEGP-ADGAPE-LPER--------SMYAAWARILGREYERLLRDEASL-------------------KRTVIDPYGATNPAEFFAVVTEAFFEKPRKLRAKHPELYEQLKAFYCQDPAS-----R---------------------
gi_149178267_ref_ZP_01856 1 --------------------------------------------MLF---------------------------TWWRNR----RRRKILA---SPMPDSWLTILD-QNVSQLSRLTEAQRTLLYQRVQIFFKEKYWEGCNGFE-ITEEVKLTIAGQACLLTLGFA-----TDCFDRLQTVLVYPDTYMARE---EMLNSIGVLTEGTSFRLGESWGQ-GPIVLSWESI-REGG-------QIPDDG--RNVVIHEFAHYYDAIDRQ-FNGTPL-LSGP--------EEYRRWNEVMTREFQRLVSQVQQG-------------------QDTFIDPYGSTNPAEFFAVCSEHFFEQPRLMQEYSVDLYETLRMFYRQDPAR-----QY--------------------
gi_77360113_ref_YP_339688 1 --------------------------MINGLLIVILI---IFVVIFWRW-------------------------PAIQNR---LWQRKYKT---LELNSQQKDILL-RCMPIYKKMTDADRAKLERHIMWFLSEKRFVGCDGLK-LTGAMKLIVAADACLLVLNKP-----WPLYKNVKEILLYPSAYYAPQ---LTRDSAGLVSYHNAIRLGESWPG-GTLVLSWHDV-LEGN-------RLPSDG--HNLVFHEFAHQLDQETGK-TTGTPL-LKTA--------AEYKEWGRVFSRAFNQLKSHVAYS-------------------MEHVIHSYGATNEAEFFAVITETFLEKPAELRRYDPEIYRLLVDYYQFDPIV-----WH--------------------
gi_163722977_ref_ZP_02130 1 --------------------------MF----------------------------------------------RWFADR----RRKKLTQ---APFPPVWKEILR-QNVAYYSMLKAEERVKLHALIQVFVAEKNWEGAGELE-LTDEIRVTIAAQACLLLLGLP-----HNYYRNVQSIIVYPSTVTPPQRKRSFFESSPTSVEAPQPILGQAFKG-GPVILCWDAG-LRGG-------RNSEDG--HNLVYHEFAHKLDMLDGA-ADGTPP-LRDR--------AEYRDWVQVCSEEFLRLKKNAKNG-------------------RKSFLRSYGAVNEAEFFAVATEQFFEQPQKMVEHAPELYRVLKEYYRQDPYR-----RASRRNNKV-------------
gi_16329696_ref_NP_440424 1 --------------------------MLPTAIVLTIL--LGIIAYIWGY-------------------------PHWLDW----REKQLFQ---RPLPPHWQAILG-DRLPFYAQLSPQQRQKLEAKIQLFLQQKQFIGCNDFV-LTDEVRLVIAAQACYLALELG-----SNPYPRLDTILVYPDAFQVRQ---ITSPDGYVVEEEDTVRAGESWDRAGQLILAWGTI-AWDL-------ERWQDG--HNVIFHEFAHQLDMGDGA-MNGVPK-LGRR--------NDYQRWQSIFASEYQQLLSQLENN-------------------LPTVIDPYGATNPCEFFAVVTETFFEKGQELQHNHGQLYQVLRKYYCLEPLI-----NC--------------------
gi_152992626_ref_YP_00135 1 -------------------------MASYYLNLLLFVSLLGLTILGYWL------------------------IKWYRQK---QELQRIAA---MPFE-EEDRSVL-SRIKQYQYLSSEEKEKIERSILRFMNTKNFIGA-KLE-VTEEMKVVISFYACLLLLHID----TENCYDNLKNIIIYNHPVMIDR----VQNNGGIFSKEQFLIDGQSAN--DTVVIIWHDA-KREA--------YHPRK--ENVIVHEFAHEIDFMDGE-IDGVPP-MER---------SKYHEWTNVLYKEFEKLNKVAMK----NRDW-----------GDYKFIGEYAATNEAEFFAVISERFFESPGSLKKKFPDLYNELKDFYGVDLAA-----KEA-------------------
gi_87310612_ref_ZP_010927 1 --------------------------------------------MFF---------------------------SWFKQR----SRDRIKS---QPFSSDWRTFVE-THLWQYPALTPAEQEKLQHSIAILVAEKNWEGCNGVQ-VDELHKVAIAAQVGLMTLGLG-----EEYFDNVMSVLIYPSAYRATS---PVASAGGVVIESQSDRLGEAWYR-GPVVLSLPDV-LEGA-------RSPNHA--RNLVVHEFAHQLDMLNGRSADGIPP-MNSD--------EQAQSWIHNFQRAYQRQVQDCQSG-------------------HRPLVDCYGATNEAEFFAVLTEAFFQAPKHLAAADHDLFMSLQSYYQQDPLR-----WTH-------------------
gi_115378754_ref_ZP_01465 1 --------------------------MS----------------------------------------------GLFRTL----RRRRLLR---RPFPAEWLGYLE-RRAPFLQKMAPEPRQRLLDLLKVFAWEKEFIGAGGFT-ITDEVRAVISATAARLVLYLD-----LSYYDRLREVIVYPDAFRIPD--------------RTGVVLGEAKNW-GTVILSWQSV-LAGL-------SDPNDG--HATATHEFAHVLDRQDGA-FDGTPH-LRRY--------SHYGAWAEVMGEHFQKLRQGQRA--------------------ERQVMDDYGSVNEAEFFAVATESFFEKPWQMKEKTPDLYEELKRFYGWDPAS-----TPP-------------------
gi_190572354_ref_YP_00197 1 --------------------------ML----------------------------------------------QWLRSA-------------PRPIDDALWDEAC-RRAAWLHGLDDDRRARLRLLATRFLHEKTITPIGELQ-LHAGDGVLLAALCCLPLLEFG-----EVGLEGWSQLIVYPDAFRVHR---SHMDAAGVLHEWDDELIGESWDS-GPLILSWADV-QADL-------ATPHEG--YCVAVHEMVHKLDALDGA-MDGTPP-LPR---------EWQREWAATFQRSYDAFCAQVDAG-------------------EETLIDPYAAEAPEEFFAVASEYHFSAPDLLQQALPEVAAQLRRFYGEPPKI-----ETR-------------------
gi_166713910_ref_ZP_02245 1 -------------------------MAQFPAG------------HVPLI-------------------------QRLTRW------LRRPP---PDIADALWQPLC-SRCAWVAALDPPRRQRLRALTAHFLHQKTISPVDGLQ-LQPSDALMLAATCCLPLLEIG-----AAGWRGWSQLIVYPDAFRVQR---THTDAAGVMHAWDDTLIGEAWEH-GPLIVSWADV-QTDL-------DDPQAG--FNVIVHEMAHKLDALDGA-LDGSPP-LPR---------AAQRTWARDFQHAYDLFCQRVDAG-------------------EDTEIDPYAAEAPEEFFAVVSEYHFSAPHLLARAMPQVAAHLERFYGPSPFA-----ASVQR-----------------
gi_21233361_ref_NP_639278 1 ----------------------------------------------------------------------------MLRW------LRRPP---AALDDALWQAVC-TECAWVTALDPPRQHRLRALTTQFLQQKTISPVHGLT-LTPHDAVLLAATCCLPLLNIG-----AAGWRGWSQLIVYPDAFRVQR---THMDAAGVMHAWDDTLIGEAWEH-GPLIVSWADV-QADL-------ADPTAG--FNVVVHEMAHKLDALDGA-LDGTPP-LPA---------AQYRDWARDFQQAYTAFCRQVDAG-------------------TQTDIDPYAAEAPEEFFAVLSEYHFSAPQVIAREMPRVAAHLTRFYGASPFA-----HAPPN-----------------gi_77461216_ref_YP_350723 1 --------------------------MW------------------------------------------SLS-AWRRR-------RILAR---HPIADELWQRVR-HHLSFLDGITAAEDQWLREASVLFLEDKHLTALPGVE-LHQEQRLLLAAQAQLPLLHLG----DLNWYQGFHEIVLYPDDFLSPQ---RHRDASGVEHEWDGEHSGEAWQQ-GPIILAWPGV-MASG---------GWEG--YNLVIHELAHKLDMLNGD-ANGLPP-LHAD--------MRVSDWAEVMQAAYDDLNRQLDRHP-----------------DAETAIDPYAAENPAEFFAVTSEYFFSAPDLLHEAYPQVYAQLQLFYCQDPLA-----RLKQLQATDPVYQAHE------
gi_146283997_ref_YP_00117 1 MAAEVRLAAGERRAYGERPAPLETSAMW------------------------------------------SFR-AWQRK-------RILAR---NPVDVDLWQRVL-DSLPILDGLGDDELARLRERAVLFLHDKRLTALPGVE-LQAEDRLRLALQAQLPLLHLA----DLGWYHGFHEIVIYPDDFLSPQ---KHRDAAGVEHEWDAEHSGEAWLQ-GPVILAWPGV-QESG---------GWEA--YNLVIHELAHKLDMLNGD-ANGLPP-LHRQ--------MRIDAWASAMQGAYDRLNAALDADP-----------------EAETPIDPYAAENPAEFFAVTSEYFFSAPDVLQQSFPDVYAQLAAFYRQDPLA-----RLQRLQAEHPAYREPA------gi_146308843_ref_YP_00118 1 --------------------------MW------------------------------------------SLK-AWRRN-------RILER---NPIDPAHWAEVC-RRLPILDGLDEHEQQRLRERAVLFLHDKHLTALPGVE-LGPVERLLLSAQAQLPLLHLG----ELDWYRGFHEIVLYPDDFVSPQ---RHRDAAGVEHEWDDPRSGEAWHQ-GPVILAWPGV-EASG---------SWEG--YNLVIHELAHKLDMLEGG-ANGLPP-LHRD--------MRVQDWANAMQSAYDALNAQLDADP-----------------DAETAIDPYAAEDPAEFFAVTSEYFFSAPDLLAEAFPEVYRQLALFYRQDPLA-----RLQRLQAEHQDYQMVENATNG-gi_70732782_ref_YP_262545 1 --------------------------MW------------------------------------------SLS-AWRRK-------RRLAR---HPVSDETWQRVR-QHLPFLDGISAVEDQWLREASVLFLDDKHLTALPGVE-LHQEQRLLLAAQAQLPLLHLG----DLNWYQGFHEIILYPDDFLSPQ---RHRDASGIEHEWDGEHSGEAWQQ-GPIILAWPGV-LASG---------GWEA--YNLVIHELAHKLDMLNGD-ANGLPP-LHSD--------MRVSEWAQVMQSAYDDLNHQLDRNP-----------------DAETAIDPYAAENPAEFFAVTSEYFFSAPDLLVAAYPKVYAQLQLFYRQNPLA-----RLQQLQAEDPVYQTHH------gi_66043892_ref_YP_233733 1 --------------------------MW------------------------------------------SFS-HWRRQ-------RTLAR---HPVAEELWQNVC-ARLPILDGLGNGQHAQLRDACVLFLHDKHLSALPGVD-LDDEQRLFLAAQAQLPLLNLG----ELNWYQGFHEIVLYPDDFVSPQ---RHRDASGVEHEWDGEHSGEAWLQ-GPVILAWPGV-LSSG---------GWDG--YNLVIHELAHKLDMLNGD-ANGLPP-LHSD--------MQVTEWARVMQSAFDDLNRQLDHNP-----------------DAEPAIDPYAAEDPAEFFAVTSEYFFSAPDLLHGSYPAVYEQLKAFYRQDTLA-----RLDLLRQQDPAYRDS-------gi_167031600_ref_YP_00166 1 --------------------------MW------------------------------------------SFS-AWRRK-------RTLAR---YPITPQQWQAVR-ERLPMLDGITDAEDQWLREACILFLLDKHLTCLPGVE-LDDEQRLLLAAQAQLPLLHLG----ELNWYQGFHEIILYPDDFKSPQ---RHRDASGVEHVWDGEHSGEAWQQ-GPIILAWNGV-LGSG---------GWEA--YNLVIHELAHKLDMLNGD-ANGLPP-LHSG--------MPVDEWATAMQQAYDDLNRQLDADP-----------------DAETAIDPYAAENPAEFFAVTSEYFFSAPDLLHQAYPQVYRQLSQFYRQDPLA-----RLCRLQAEHPEYRESHA-----gi_16121989_ref_NP_405302 1 --------------------------MI----------------------------------------------KWLWK-------ANKPQ---AEM-LAQWHEAL--NIPLLAPLNEPEQQRLVSVASQLLQQKRFIPLQGLI-LTPLMQARLALLFALPVMELG-----AKWLDGFHEVLIYPSPFIVAE---DWQDDLGLVHSGQSVQSGQSWEQ-GPIVLNWQDI-QDSF---------DLSG--FNLVIHEAAHKLDMRNGGHSNGVPP-IAM---------RDVAVWEHDLHHAMDNIQDEIDMVG-----------------VEGASMDAYAASNPAECFAVLSEYFFSAPELLEGRFPAVYQHFCRFYRQDPLA-----RLKRWENSLADNPPPENTHSHR
gi_123442833_ref_YP_00100 1 --------------------------MI----------------------------------------------KWLWK-------ANQPQ---AEM-LAQWREAL--NIPLLAPLNEEEQQRLVSVASHLLQQKRLVPLQGVV-LTPLMQARLTLLFALPVMELG-----AKWLDGFHEVLIYPSPFVVDD---DWQDDAGLVHSGQTVQSGQSWEQ-GPIVLNWQDI-QDSF---------DLSG--FNLVIHEAAHKLDMRNGGHSNGVPP-IAM---------RDVAAWEYDLHQAMDNIQDEIDMVG-----------------VDAASMDAYAASDPAECFAVLSEYFFSAPELLENRFPLVYRHFCTFYRQDPLA-----RLKRWENESQNIESSSPMG---T0489_Putative_Metal-depe 1 -------------------------MMF----------------------------------------------KWPWK-------ADDES---GNA-EMPWEQAL--AIPVLAHLSSTEQHKLTQMAARFLQQKRLVALQGLE-LTPLHQARIAMLFCLPVLELG-----IEWLDGFHEVLIYPAPFIVDD---EWEDDIGLVHNQRVVQSGQSWQQ-GPVVLNWLDI-QDSF---------DASG--FNLVVHEVAHKLDTRNGDRASGVPL-IPL---------REVAGWEHDLHAAMNNIQDEIDLVG-----------------ESAASIDAYAATDPAECFAVLSEYFFSAPELFAPRFPALWQRFCHFYRQDPLA-------RRRENGLQDEGDRRIVH---gi_119897976_ref_YP_93318 1 --------------------------ML----------------------------------------------EKLGRW---LTERFGRT--PPEVAEADWQQAE-AGLPFLAYLDDADRLRLRELARRFLAEKQFHGAHGLQ-LTDHMLLTISLQACLPVLNIG-----LSAYRGWVGVVVYPGDFVIPR---QEVDEAGVVHEYDDEVLGEAWSG-GPVLLSWFDE-G-A-----------PEG--VNVVIHEFAHKLDMENGG-VDGLPV-LPPS--------MSRSAWAQVFGEAYERFCDQVDHG-------------------IDSLLDPYAAEHPAEFFAVAAEAFFEMPLDLRDEFPAVYDQLARYFRLDPAH-----RASSLLLTPDPTSFPPP-----
gi_56479170_ref_YP_160759 1 --------------------------ML----------------------------------------------NFLRKW---RKARRLAA---IRVPDALWTSVE-TKLPFLDYLESTDRERLRQLAREFLAEKQFHGAHGMA-LTDQMMLTIALQACLPILCVG-----LDAYRGWVGVIVYPGDFVIPR---HETDDAGVVHEYDDEVLGEAWEG-GPVLVSWHRE-GEG-----------PEG--VNVVIHEFAHKLDMQNGD-VDGLPL-LPQG--------MSRAAWAKVFTDAYQRFCEQVDAG-------------------EETLLDPYGADSPGEFFAVASEAFFETPCLFEQELPRVYEQLSRFYRLDLAAGERRVRDKRRCHDGRPLRTFGS-----gi_169182115_ref_ZP_02842 1 --------------------------ML----------------------------------------------QLFRRL---LGIPPAR----PLVDESQWRRVE-ARLPFLDHLVPDDRLRLREMALAFLGCKEFHGAQGLE-LDDDMLLAIALQACLPVLHIG-----LQAYEGWVGVIVYPGDFLIPR---RIVDEAGVLHEYEDEVLGEAWAG-GPVLLSWFPG-NDA-----------PAG--VNVVIHEFAHKLDMENGD-VDGLPR-LRPG--------MSRRDWAKAFGEAYEAFCAEVDRG-------------------IDTTIDPYGAEHPGEFFAVVSEVFFEDPLVLHRRFPAVYDQLVHFYGMDPAA-----RAAA-----AAGG---------gi_187928120_ref_YP_00189 1 -------------------------MLSSVF-------------------------------------------RWFSGR---ARARRLTR---YAISDARWSRAL-DGLPFLLDWPGTDLARLRETATLFIAEKEFTTAHDLP-LTDDMVVSIAAQASVPILELG-----ITWYSGWHGVVIYPGEFVIRK---EVMDEDGVVHNVREEAAGEAWEH-GPVILSWQDI-ELGGA----LAEPGAQP--YNVVVHEFAHKLDMLNGE-SDGIPA-FSSRLH----ANVDREQWADDLYAEYDAFANRCEQLPERRWGE----------DPILSLLDPYGAQHPAEFFAVASEAFFVEPLALLETLPSLYALLQAFYLQDPAR-----RVLDSEQRQEGFLP--------
gi_113868370_ref_YP_72685 1 -------------------------MLGKLT-------------------------------------------SFLRTR---SRARKLEH---YAIPDALWARTV-ATLPFVQRYGSEDLAALRELATLFIADKEYSTAHDLP-LADDMVASVAVQACVPILKLG-----LPWYRGWHGIVLYPGEFIIRR---TVQDEIGLVHGVVEEASGEAWEH-GPVILSWPDV-HTPGAGIDSDHELPADT--YNVVIHEFAHKLDMLDGE-PDGVPP-FSRVLH----PDIDAEAWAETFLTEYDRFAESWEARDEADWEHPE------RLPPALRIIDPYGTEAPSEFFAVTSEAFFVEPTGLARHWPVVYQSLAAFYRQDPAS-----M---------------------gi_73541807_ref_YP_296327 1 -------------------------MLGKLT-------------------------------------------QFLRTR---SKARKLEH---YAIPDALWDHTV-SALPFVQRYPEDDRRALRELATLFIAEKEYSTAHELP-LSDDMVVSVAVQACVPILRLG-----IEWYRGWHGIVLYPGEFIIRR---TVEDEIGLVHGIVEEASGEAWEH-GPVILSWPDV-ESPGGGLDTEHDVPADT--YNVVIHEFAHKLDMLDGE-PDGVPP-FSRVLH----PGVDAEAWAETFLTEYDRFAETWDRHPVEAWEHPE------RLPPALRLIDPYGCEAPSEFFAVTAEAFFVEPVGLKRHWPVVYQCLATFFRQDPA--------------GTSA----------gi_17545903_ref_NP_519305 1 -------------------------MLSRLS-------------------------------------------QWLSRR---ARSRQLAR---YAISDALWTRTL-SGLPFLLDWPQAALARLRETATLFIAEKEFTTAHGLA-LTDEMVVSIAAQASVPILELG-----IAWYRGWRGVVLYPGEFLIRG---EAMDEDGVVHDVRQEASGEAAAN-GLVLLSWQDI-ELGSV----LAGPDMQP--YNVVMHEFTHKLDMLNGE-ADGIPA-FSSRLH----AGLDREQWADDLYAEYDAFAERCDRIPERRWDA----------DPILSLLDPYGAQHPAEFFAVASEVFFVEPAALQDTLPALYALLQAFYLQDPAR-----RMLDAGRHAEPATP--------gi_161524614_ref_YP_00157 1 -------------------------MLSKLT-------------------------------------------RWLDDR---RRDRALRS---HPIPDTLWRDTV-ARLPFLDALSPDDLGRLRELTSLFIAKKSFSTAHGLE-LTDAMIVAIAAQACLPVLNLD-----LSLYDGWVGVVVYPGEFVIRK---TVQDEDGVVHEVEQDASGEAWEG-GPVILSWEDA-QMTD---------GRDA--YNVVIHEFAHKIDMVNGA-ADGYPP-LFRRWHA---PQLDAQIWADVFDNAYDQFCARVDAVPDRAWAR----------FERDSLIDPYAADHPSEFFAVCSEALFVRPQAFESEFPELYRLLARYYRQDPAR-----AGALDAA---------------
gi_167719238_ref_ZP_02402 1 -------------------------MLSKLN-------------------------------------------QWLDAR---RRDRALRS---HPIPDVLWRETL-ERLPLLSYLSDEDRARLRETTSLFLAQKSFSTAHGLE-LTDAIVVGIAAQACVPVLNLG-----LDLYRGWVGIVVYPGEFVIRK---TVEDEDGVVHEVEHDASGEAWEG-GPVILSWEDV-QMAD---------GSDA--YNVVIHEFAHKIDMMSGA-ADGHPP-LFRRWHA---PALDAERWADVFDHAYDQFCDRVDAVPERAWAR----------FERDSLIDPYAADHPSEFFAVCSEALFVKPRPFEAEFPELYRLLARYYRQDPAR-----TGCLAGSAAHV-----------gi_83720796_ref_YP_442735 1 -------------------------MLSKLN-------------------------------------------QWLDAR---RRDRALRA---HPIPDALWRETL-ERLPFLSYLSGEDRERLRETTSLFLAQKSFSTAHGLE-LTDAIVVGIAAQACVPVLNLG-----LELYRGWVGIVVYPGEFVIRK---TVEDEDGVMHEVEHDASGEAWEG-GPVILSWEDV-QMAN---------GSDA--YNVVIHEFAHKIDMMSGA-ADGHPP-LFRRWHA---PELDAERWADVFDHAYDQFCDRVDAVPERAWAR----------FERDSLIDPYAADHPSEFFAVCSEALFVKPRPFEAEFPELYRLLARYYRQDPAQ-----TGCLAHHAAHV-----------gi_53723587_ref_YP_103027 1 -------------------------MLSKLN-------------------------------------------QWLDAR---RRDRALRS---HPIPDVLWRETL-ERLPFLSYLSNEDRARLRETTSLFLAQKSFSTAHGLE-LTDAIVVGIAAQACVPVLNLG-----LDLYRGWVGIVVYPGEFVIRK---TVEDEDGVVHEVEHDASGEAWDG-GPVILSWEDV-QMAD---------GSDA--YNVVIHEFAHKIDMMSGA-ADGHPP-LFRRWHA---PALDAERWADVFDHAYDQFCDRVDAVPERAWAR----------FERDSLIYPYAADHPSEFFAVCSEALFVKPRSFEAEFPELYRLLARYYRQDPAR-----TGCLAGSAAHV-----------gi_134295863_ref_YP_00111 1 -------------------------MLSKLT-------------------------------------------RWFDDR---RRDRALRS---HPIPDALWDDTV-ARLPFLGALAPDDQRRLRELTSLFIARKSFSTAHGLE-LTDRMIVAIAAQACLPVLNLD-----LSLYDGWVGIVVYPGEFVIRK---TVEDENGVVHEVEQDASGEAWEG-GPVILSWEDA-QMTD---------GRDA--YNVVIHEFAHKIDMLTGA-ADGYPP-LFRRWHA---PHLDAQAWADVFEHAYDQFCARVDAVPERAWAR----------FERDSLIDPYAADHPSEFFAVCSEALFVRPQAFESEFPELYRLLARYYRQDPAG-----TGALDA----------------gi_167587024_ref_ZP_02379 1 -------------------------MLSKLT-------------------------------------------RWLDHR---RRDRALRS---HPIPDALWQDTV-ARLPFLDYLPPDDLGRLRELTSLFIAHKSFSTAHGLE-LTDEMIVGIAVQACVPVLNLD-----LSLYDGWVGVVVYPGEFVIRK---TVQDEDGVVHEVEQDASGEAWEG-GPVILSWEDA-QMTD---------GRDA--YNVVIHEFAHKIDMINGA-ADGYPP-LFRRWHA---PHLDAQAWADVFENAYDQFCARVDAVPDRAWAR----------FERESLIDPYAADHPSEFFAVCSEALFVRPRAFEAEFPELYRLLARYYRQDPAQ-----TGALDAR---------------gi_33594455_ref_NP_882099 1 --------------------------ML----------------------------------------------RWLRGRGAKAGEVAAMQ---ARIPPDLWRRTL-HAYPFLAALSEAEQQALQARAAWLLASKNINGAHGLE-ITDFMRLSVAAQAALPVLNLS-----PTLYEGWDEIIIYPAGFSIQR---ERMDEDGVMHEYIEEAVGEAWEG-GPVILSWSDA-AGS-----------DGA--FNVVIHEFAHKLDLLAGQ-ADGMPA-LAAHP------DIDPRQWRRVLEDSLDRFAAAVEAVERAIPAHVDPESPQADPWYARLPLDPYAATDEAEFFAVSSEHFFVEPEPLADALPQWYALLRAYYRQDPLA-----RLA-------------------
gi_187477598_ref_YP_78562 1 --------------------------MF----------------------------------------------RWLTGRGASQAEVQEMQ---ARIPEPLWRDVL-AAHPFFDALDAADLEALRARAAWLLASKQINAAGCLE-LNDFMRLSICAQAALPILKLS-----PQLYAGWDEIIVYPSGFTIPQ---VMQDEDGVVHEYLEEAAGQAWDG-GPVILSWDDA-ADA-----------QEA--FNVVIHEFAHKLDLRAGS-ADGMPD-LGPG--------LSPRQWQQTLQDSLAHFRRALDEIEAAIPPDIDPESPDADTWYGQLPLDPYAATDEAEFFAVSSEHFFTAPWPLAEAMPDWYALLRAYYQQDPLA-----RLS-------------------gi_88811628_ref_ZP_011268 1 --------------------------MT----------------------------------------------GWLRRW---RRKRILAQ---KRIPERLWQEVI-AAQPLLMRMSVPAQQQLHELATLLVAEKRFYGIAGLQ-VGPWQTTTIAALGCVPVLELG-----LDWYRGWCSIVIYPGGFRARH---AYPDDAGVIHEQVTELIGEAWDG-GPLVLSWEDI-QGGV---------RLDG--YNVVLHEFAHKLDMRNGV-ANGMPP-LHRG--------MSRRDWTRAFTNAWNQAQAAAAAG-------------------EELVLDRYALESPGEFFAVACEVFFELPERLCAGYPAVYEQLRRFFRQDPLQ-----RSSSSQAIYRPLTTERN-----
gi_117618776_ref_YP_85659 1 --------------------------MFW---------------------------------------------SRLRSV---FGGASRQD--GNGPGALEWQQAL-AAVPILQGFSREEEQTLIGLAQQLLARKTLTQL-GVS-LTSRQQAVLALQAALPILNLG-----LDWYRGFHEIILIPEPVVRRQ---QVQDEFGLVSEVEEENAGESWPQ-GPLVLAWSEL-QQDG---------DWDG--FNLVIHELTHKLDMLGGE-ADGAPP-LPAG--------MDPRQWSGALQAAFDAMQGQLDRH-------------------EPTAIDPYAASHPAEFFAVCSECFFTDPLRLRDTYPAVYQQFTRFFGQQPAA-----RFPATRLLAGEPEYQG------
gi_94264888_ref_ZP_012886 1 --------------------------MW----------------------------------------------RWLAAW---WRRRLLRR---AALPDELWRQAVARPRELFGGLDYDEQRRLRELATLLLARKRFYGGGDLV-VTPFMRASIAAQACVPILNLG-----LACYDGWTSIVVYEHSFVVPR---EEVDEDGIVHSGNEVLSGESWEA-GPLVLSWAEC-DPGA-------HPHGSA--GNVVIHEFAHKLDMLSGA-VNGTPA-LPGH--------LDANLWAATMEAAWQSLHAARDRG-------------------EELPVDEYATQDPGEFFAVLSEYFFMAPAVLYRAFPEVYRQLALYYRRDPLA---------------------------
gi_121996823_ref_YP_00100 1 -------------------------------------------------------------------------MGWLRNW---RRQRIAQR---YRLSPAAWSRAE-AALPGLRRLPPGTRQRLAAAATALIHEKRFEGVAGVT-LDALAVDTVALQAAVPVLELG-----VDWYAPWTTILLYPAGFVATH---EYEDEDGVVHVEQGPLIGEAWEG-GPLVLSLEDA-LAP-----------EPG--SSVVIHECAHKLDMGQGA-VNGLPP-LPRG--------MTVDDWSAAFLPAFHTMEAYAAEGA-----------------CAVTPLDPYAGQDPGEFFAVATETFFADPHRLRRAFPEVYRQLARFYGWSLG----------------------------
gi_114320551_ref_YP_74223 1 --------------------------MF----------------------------------------------RLDRLW---RGSRPG-----EPLNNLHWHEAM-ARCWPLPALSADERPQLEALAREFLASRAWEGVGGLD-PDERLRLAVTVQACLPVLSLG-----LDWYSDWVTVLLYPSGFVARH---VWEDEFGVVHEEEGPLVGEAWER-GPLVLSAEDV-LTP-----------EPG--FSVVVHECAHKLDMRNGD-ANGQPP-LHAE--------MSPARWKAAFSAAWDRFAEGVEAQA------------------DWLPFDAYAAENPGEFFAVASEAFFTDPAPVQQVFPEVYQQMVAFYRQDPLA-----RLTRAGLLP-------------
gi_93006198_ref_YP_580635 1 --------------------------MF----------------------------------------------SMIKDW---REQRILDN---SEFTHADWMQAA-KRIVILDRLNEDELTRLFELATLFLADKSITGAQGFE-ITDAVKQSIALQACLPILKLS-----LEWYAGWSAIIIYPGSYKSET---TTVDELGIVHEGSQHRSGEAWLR-GPVILSWKDA-KHSG---------ERDG--HNVVIHEFVHKLDMLNGR-ANGFPP-LQPD--------MDPARWTEIMSRDFENFQSHHK-----------------------SGLDRYGATNPAEFFAVLSEVFFETPQKLIDAYPDIYEIMVKFFRQTPLE-----PRP-------------------
gi_71065647_ref_YP_264374 1 --------------------------MF----------------------------------------------SMTKDW---REQRILDS---SEFTHADWLQAA-QRIVILGRLNEDELTRLFELATLFLADKSITGAQGFE-ITDAVKQSIALQACLPILNLS-----LEWYAGWSAIIIYPGSYKSET---TTVDELGIVHEGSQHRSGEAWLR-GPVILSWKDA-KHSG---------ERDG--HNVVIHEFVHKLDMLNGR-ANGFPP-LQPD--------MDPVRWTEIMTRDFEDFQSHHK-----------------------SGFDRYGATNPAEFFAVLSEVFFETPQKLIDAYPDIYEIMVKFFRQTPLE-----PRP-------------------gi_53804966_ref_YP_113359 1 -------------------------MDDTATG------------QTLPI------------------------LKFFRHL---RRRRILDE---CPIGPTAFRAAI-ANVPLLARLPEPEREKLRRLATLFLHEKIFESSGNLA-LTEAMKIRIAALACLPILHLD-----LDWYAGWRTVVVFPTEFVRPR---EAMDDVGVMHEWEEILGGEAWDR-GPVILSWADV-EASG---------QGDG--YNVVIHEMAHKLDMLNGP-CDGFPP-LHRG--------MRPSAWTQDFTAAFEDFNVRLDRA-------------------EYTPVDPYAGESPSEFFAVLSEYFFERPALVHDEFPAVYAQMCQFYRTDPLQ-----RTPSLG----------------
gi_160897943_ref_YP_00156 1 --------------------MSLWQQMG----------------------------------------------SWVRRL---GRRARSTVSPVPEIPAPLWLSTL-AAYPFLASLTLEEQSKLRALAAVFLQRKRFHGAHGLQ-VSDAMALEIAAQACLPLLHWGEPQGALSWYDDFVGIVVHPGDAVARR---QSMDEAGVIHDYDEVIMGEAMHG-GPVMLSWQAI-GAGRGSHASHHGHGGQP--ANVVIHEFAHKIDMRNGG-ADGCPP-LPPGFMGSSSARQAREIWQAAWAPAYDAFREQITIA---ERFG-----------GEWPWLDGYGATAPAEFFAVACEAFFVDRARFALEFPTLAPLLAALFKPATSQ-----P---------------------
gi_121593991_ref_YP_98588 1 -------------------------MARTRGPL-----------------------------------------HFLRQL---ALRVRATVAPLPDISAQLWLQTL-RRYPFLQALPLHDQGKLRALSALFLDRKEFTGAHGLV-VTDAMAVSIAAQACLPLLHWGAPERALAWYDDFVGIVVHPAEAVARR---QAVDAAGVVHHYDEVLLGEAMER-GPVMLSWPAVDGAGQ------GGPDGQPVATSVVIHEFVHKIDMRNGP-ADGCPP-LPPGFMGTRDARAAHAAWRDAWEPAYEQFREQVIIA---ERFG-----------GAWPWLDAYGATAPAEFFAVACEAYFVHRERFGQEFPTLLPVLDAFFGRPLSG---------------------------gi_171058348_ref_YP_00179 1 --------------------------------------------MLS---------------------------RWWRKQ---RDARALRQ---RAIPDELWLDTL-RRHPFLTLRPLDDLLKLRSLCALFLDRKEFTGADGLE-VTDEMAVAVAAQACLPVLHLD-----LAAYDGFVGIVMHPGEVVAQR---EWIDEDGICHQGEEELAGEAMPG-GPVMLAWSEV-DAGG-------ELAASG--YNVVIHEFVHVLDMLDGE-ADGVPP-LPAR--------ADRAQWLAVIEPAFERLRQQVEAD-------------------EDTMLDPYGASAIEEFFSVASEAFFVAPDLLIATEPAVYRLLRDYFRQDPAR-----YAGIRPR---------------
gi_92114829_ref_YP_574757 1 -------------------------------------------------------------------------MKWWNA-------WRGAR---HPFPAEAWAEGL-ARVPLLQALPAEEANRLGERAWAFLRRKRLALFDDEL-FDLPARLALVAQACLMTLHWSED-EAREAFANVHEVRVLPEAFRRRV---EEMDEAGVVHEYEDERVGETWHQ-GPIVLALSDI-QESG---------GWGG--FNVVIHEFAHKLDMGNSLDADGFPP-LPRD--------IDARDWHRVFTGVWDDLQAHLARG-------------------DAPPIDDYAASHPGECFAVCCEYFFTAPDILDAAYPELYALLARYFRQAPLQ-----RCPPH-----------------
gi_110833977_ref_YP_69283 1 --------------------------MF----------------------------------------------AAWKNW---RDARRVKK---MGYTREEWEAAV-ADWPLMVRFEGQERQALFDLSLRFLARKSIASGGGFA-YTDAMCLKVATMACVPILYLG-----LDWYDGWYTVILYEDAFIPNR---PHFSEDGVVHTRGPVLAGEAWHQ-GPVILSWDAI-EEAC-------APSNQA--SNVVIHEMSHKLDMRRNG-ANGRPP-LHTG--------MNPKQWHETFTACWEELLSDYQHH-------------------RHMPVDAYALTGPGEFFAVCSEAFFEGPDQLYKDWPALYRLLAQFYRQGER----------------------------
gi_16121989_ref_NP_405302 1 --------------------------MI----------------------------------------------KWLWK-------ANKPQ---AEM-LAQWHEAL--NIPLLAPLNEPEQQRLVSVASQLLQQKRFIPLQGLI-LTPLMQARLALLFALPVMELG-----AKWLDGFHEVLIYPSPFIVAE---DWQDDLGLVHSGQSVQSGQSWEQ-GPIVLNWQDI-QDSF---------DLSG--FNLVIHEAAHKLDMRNGGHSNGVPP-IAM---------RDVAVWEHDLHHAMDNIQDEIDMVG-----------------VEGASMDAYAASNPAECFAVLSEYFFSAPELLEGRFPAVYQHFCRFYRQDPLA-----RLKRWENSLADNPPPENTHSHR
gi_123442833_ref_YP_00100 1 --------------------------MI----------------------------------------------KWLWK-------ANQPQ---AEM-LAQWREAL--NIPLLAPLNEEEQQRLVSVASHLLQQKRLVPLQGVV-LTPLMQARLTLLFALPVMELG-----AKWLDGFHEVLIYPSPFVVDD---DWQDDAGLVHSGQTVQSGQSWEQ-GPIVLNWQDI-QDSF---------DLSG--FNLVIHEAAHKLDMRNGGHSNGVPP-IAM---------RDVAAWEYDLHQAMDNIQDEIDMVG-----------------VDAASMDAYAASDPAECFAVLSEYFFSAPELLENRFPLVYRHFCTFYRQDPLA-----RLKRWENESQNIESSSPMG---T0489_Putative_Metal-depe 1 -------------------------MMF----------------------------------------------KWPWK-------ADDES---GNA-EMPWEQAL--AIPVLAHLSSTEQHKLTQMAARFLQQKRLVALQGLE-LTPLHQARIAMLFCLPVLELG-----IEWLDGFHEVLIYPAPFIVDD---EWEDDIGLVHNQRVVQSGQSWQQ-GPVVLNWLDI-QDSF---------DASG--FNLVVHEVAHKLDTRNGDRASGVPL-IPL---------REVAGWEHDLHAAMNNIQDEIDLVG-----------------ESAASIDAYAATDPAECFAVLSEYFFSAPELFAPRFPALWQRFCHFYRQDPLA-------RRRENGLQDEGDRRIVH---
Consensus_aa: ..........................M................................................hb...................hs......h...ph.hh..Ls..pp..h...h..Fl.pKph.s..sh..lsp.h.l.lthpthl.hh.h........hh..h..lll@Ps.h.......p...p.slhh..p....Gps....s.llltW.sh...s............ss...NlslHEhhH.lDb.s....sGhP..h................W.phh...hpph...h..........................hc.Ythps..EFFAVhsEhFFp.P..h...hP.lY..l..h@..s..............................Consensus_ss: hhhhhh hhhhh hhhhhhhh hh hhhh hhhhhhhhhhhhhhhh eeee hhhhhhhhhhhhhhhh hhhh eeee ee e ee h eeeeehhhh h hhhhhhhhhhhhhh hhhhhhhhhhhhhhhhhhhh hhhhhhhhhhhhh hhhhhhhhhhhhhhhhhhh hhh h
Target sequence - PDB file inconsistencies:
T0489 3dl1.pdb T0489.pdb PyMOL PyMOL of domains
T0489 1 MMFKWPWKADDESGNAEMPWEQALAIPVLAHLSSTEQHKLTQMAARFLQQKRLVALQGLELTPLHQARIAMLFCLPVLELGIEWLDGFHEVLIYPAPFIVDDEWEDDIGLVHNQRVVQSGQSWQQGPVVLNWLDIQDSFDASGFNLVVHEVAHKLDTRNGDRASGVPLIPLREVAGWEHDLHAAMNNIQDEIDLVGESAASIDAYAATDPAECFAVLSEYFFSAPELFAPRFPALWQRFCHFYRQDPLARRRENGLQDEGDRRIVH 266 ~~~~~~~~~~~~~~~~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~~~~~~~~~~~~~~~~|||||||||||||||||||||||||||||||||||~~|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~~~~ 3dl1A 17 ----------------EMPWEQALAIPVLAHLSSTEQHKLTQMAARFLQQKRLVALQGLELTPLHQARIAMLFCLPVLELGIEWLDGFHEVLIYPAPF-------------------------QQGPVVLNWLDIQDSFDASGFNLVVHEVAHKLDTR--DRASGVPLIPLREVAGWEHDLHAAMNNIQDEIDLVGESAASIDAYAATDPAECFAVLSEYFFSAPELFAPRFPALWQRFCHFYRQDPLARRRE------------- 253
Residue change log: change 18, 43, 71, 185, MSE to MET;
Single domain protein: target 17-98, 124-158, 161-253 ; pdb 17-98, 124-158, 161-253
Sequence classification:
DUF980 in Pfam.
Server predictions:
T0489:pdb 17-253:seq 17-253:FR; alignment
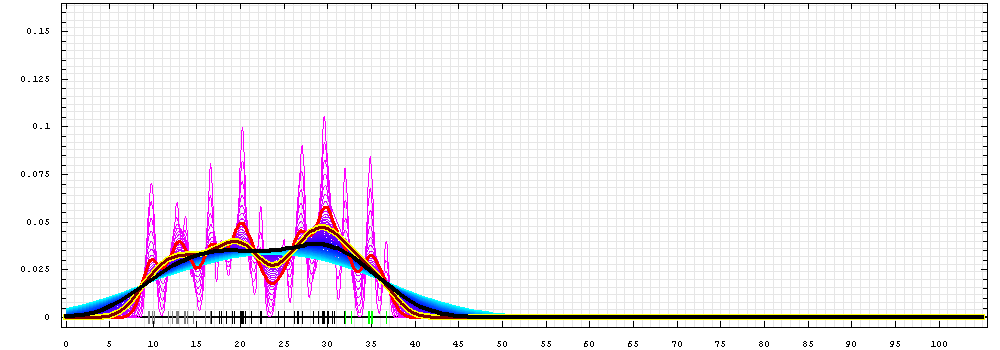
First models for T0489:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0489 | T0489 | T0489 | T0489 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | IBT_LT | 38.69 | 32.02 | 34.38 | 1.81 | 1.77 | 0.82 | 38.69 | 32.02 | 34.38 | 1.60 | 1.56 | 0.51 | IBT_LT | 1 |
| 2 | Chicken_George | 37.98 | 32.02 | 43.17 | 1.71 | 1.77 | 1.93 | 37.98 | 32.02 | 43.53 | 1.50 | 1.56 | 1.70 | Chicken_George | 2 |
| 3 | Zhang | 37.14 | 23.69 | 47.79 | 1.60 | 0.39 | 2.51 | 37.26 | 30.44 | 47.79 | 1.40 | 1.29 | 2.25 | Zhang | 3 |
| 4 | McGuffin | 37.02 | 29.76 | 43.45 | 1.58 | 1.39 | 1.96 | 38.45 | 31.43 | 43.45 | 1.57 | 1.46 | 1.69 | McGuffin | 4 |
| 5 | fams−ace2 | 37.02 | 26.94 | 41.16 | 1.58 | 0.93 | 1.67 | 37.02 | 29.29 | 41.16 | 1.36 | 1.08 | 1.39 | fams−ace2 | 5 |
| 6 | MULTICOM | 36.79 | 27.34 | 40.66 | 1.55 | 0.99 | 1.61 | 40.12 | 29.64 | 44.52 | 1.81 | 1.15 | 1.82 | MULTICOM | 6 |
| 7 | DBAKER | 36.67 | 31.91 | 40.88 | 1.54 | 1.75 | 1.64 | 37.02 | 31.91 | 43.87 | 1.36 | 1.54 | 1.74 | DBAKER | 7 |
| 8 | Zhang−Server | 36.67 | 27.14 | 39.00 | 1.54 | 0.96 | 1.40 | 38.45 | 30.36 | 43.45 | 1.57 | 1.27 | 1.69 | Zhang−Server | 8 |
| 9 | LevittGroup | 36.55 | 28.37 | 40.11 | 1.52 | 1.16 | 1.54 | 36.67 | 28.49 | 40.41 | 1.31 | 0.94 | 1.29 | LevittGroup | 9 |
| 10 | KudlatyPredHuman | 35.24 | 27.58 | 41.43 | 1.35 | 1.03 | 1.71 | 35.24 | 27.58 | 41.43 | 1.11 | 0.79 | 1.42 | KudlatyPredHuman | 10 |
| 11 | CpHModels | 35.12 | 31.11 | 28.13 | 1.33 | 1.62 | 0.03 | 35.12 | 31.11 | 28.13 | 1.09 | 1.40 | CpHModels | 11 | |
| 12 | SAM−T08−server | 35.00 | 31.43 | 40.87 | 1.31 | 1.67 | 1.64 | 35.00 | 31.43 | 40.98 | 1.08 | 1.46 | 1.37 | SAM−T08−server | 12 |
| 13 | SAM−T08−human | 35.00 | 29.21 | 39.95 | 1.31 | 1.30 | 1.52 | 35.00 | 30.28 | 40.27 | 1.08 | 1.26 | 1.27 | SAM−T08−human | 13 |
| 14 | COMA−M | 34.76 | 30.56 | 28.49 | 1.28 | 1.53 | 0.08 | 35.83 | 31.35 | 29.07 | 1.19 | 1.44 | COMA−M | 14 | |
| 15 | COMA | 34.76 | 30.56 | 28.49 | 1.28 | 1.53 | 0.08 | 35.59 | 31.31 | 29.07 | 1.16 | 1.44 | COMA | 15 | |
| 16 | METATASSER | 34.64 | 30.44 | 36.31 | 1.26 | 1.51 | 1.06 | 39.88 | 30.44 | 43.59 | 1.77 | 1.29 | 1.70 | METATASSER | 16 |
| 17 | Jones−UCL | 34.64 | 28.77 | 26.87 | 1.26 | 1.23 | 34.64 | 28.77 | 26.87 | 1.02 | 0.99 | Jones−UCL | 17 | ||
| 18 | FAMS−multi | 34.05 | 30.40 | 31.64 | 1.19 | 1.50 | 0.47 | 35.24 | 30.40 | 31.64 | 1.11 | 1.28 | 0.16 | FAMS−multi | 18 |
| 19 | SAMUDRALA | 33.57 | 28.89 | 41.08 | 1.12 | 1.25 | 1.66 | 34.41 | 29.88 | 42.70 | 0.99 | 1.19 | 1.59 | SAMUDRALA | 19 |
| 20 | GeneSilico | 32.86 | 26.86 | 40.58 | 1.03 | 0.91 | 1.60 | 36.07 | 28.77 | 43.33 | 1.23 | 0.99 | 1.67 | GeneSilico | 20 |
| 21 | LEE | 32.86 | 25.48 | 38.64 | 1.03 | 0.69 | 1.35 | 32.86 | 25.83 | 39.01 | 0.77 | 0.48 | 1.11 | LEE | 21 |
| 22 | MUSTER | 32.74 | 29.88 | 28.08 | 1.01 | 1.41 | 0.02 | 33.57 | 30.08 | 29.33 | 0.87 | 1.22 | MUSTER | 22 | |
| 23 | 3DShot1 | 32.14 | 25.24 | 27.13 | 0.93 | 0.65 | 32.14 | 25.24 | 27.13 | 0.67 | 0.38 | 3DShot1 | 23 | ||
| 24 | FFASsuboptimal | 32.02 | 29.25 | 25.55 | 0.91 | 1.31 | 32.02 | 29.25 | 30.09 | 0.65 | 1.08 | FFASsuboptimal | 24 | ||
| 25 | HHpred2 | 32.02 | 27.66 | 24.25 | 0.91 | 1.05 | 32.02 | 27.66 | 24.25 | 0.65 | 0.80 | HHpred2 | 25 | ||
| 26 | EB_AMU_Physics | 32.02 | 26.98 | 29.83 | 0.91 | 0.93 | 0.24 | 32.02 | 26.98 | 29.83 | 0.65 | 0.68 | EB_AMU_Physics | 26 | |
| 27 | HHpred4 | 31.91 | 25.00 | 24.68 | 0.90 | 0.61 | 31.91 | 25.00 | 24.68 | 0.64 | 0.33 | HHpred4 | 27 | ||
| 28 | HHpred5 | 31.91 | 25.00 | 24.68 | 0.90 | 0.61 | 31.91 | 25.00 | 24.68 | 0.64 | 0.33 | HHpred5 | 28 | ||
| 29 | TASSER | 31.31 | 26.23 | 36.73 | 0.82 | 0.81 | 1.11 | 36.31 | 29.09 | 42.18 | 1.26 | 1.05 | 1.52 | TASSER | 29 |
| 30 | Elofsson | 31.19 | 28.02 | 27.46 | 0.80 | 1.11 | 33.33 | 29.41 | 31.43 | 0.84 | 1.11 | 0.13 | Elofsson | 30 | |
| 31 | pipe_int | 30.71 | 27.18 | 29.23 | 0.74 | 0.97 | 0.17 | 30.71 | 27.18 | 29.23 | 0.46 | 0.72 | pipe_int | 31 | |
| 32 | Pcons_multi | 30.48 | 25.95 | 28.78 | 0.71 | 0.76 | 0.11 | 30.48 | 25.95 | 28.78 | 0.43 | 0.50 | Pcons_multi | 32 | |
| 33 | keasar | 30.36 | 26.07 | 29.62 | 0.69 | 0.78 | 0.22 | 33.33 | 27.86 | 40.87 | 0.84 | 0.83 | 1.35 | keasar | 33 |
| 34 | FFASstandard | 30.12 | 26.63 | 28.12 | 0.66 | 0.88 | 0.03 | 30.12 | 26.63 | 29.35 | 0.38 | 0.62 | FFASstandard | 34 | |
| 35 | FFASflextemplate | 30.00 | 25.48 | 23.06 | 0.64 | 0.69 | 30.24 | 25.64 | 24.10 | 0.40 | 0.45 | FFASflextemplate | 35 | ||
| 36 | Phragment | 29.64 | 27.10 | 32.23 | 0.59 | 0.95 | 0.55 | 32.26 | 28.14 | 34.66 | 0.69 | 0.88 | 0.55 | Phragment | 36 |
| 37 | Phyre2 | 29.64 | 26.71 | 28.67 | 0.59 | 0.89 | 0.10 | 30.36 | 27.34 | 30.59 | 0.41 | 0.74 | 0.02 | Phyre2 | 37 |
| 38 | Phyre_de_novo | 29.52 | 26.91 | 29.10 | 0.58 | 0.92 | 0.15 | 29.88 | 27.10 | 31.36 | 0.35 | 0.70 | 0.12 | Phyre_de_novo | 38 |
| 39 | pro−sp3−TASSER | 29.41 | 27.10 | 27.27 | 0.56 | 0.95 | 29.41 | 27.10 | 38.78 | 0.28 | 0.70 | 1.08 | pro−sp3−TASSER | 39 | |
| 40 | Frankenstein | 29.41 | 26.31 | 29.59 | 0.56 | 0.82 | 0.21 | 29.41 | 26.31 | 29.59 | 0.28 | 0.56 | Frankenstein | 40 | |
| 41 | Sternberg | 29.41 | 22.90 | 38.07 | 0.56 | 0.26 | 1.28 | 29.41 | 22.90 | 38.70 | 0.28 | 1.07 | Sternberg | 41 | |
| 42 | A−TASSER | 28.93 | 26.47 | 25.11 | 0.50 | 0.85 | 29.17 | 26.47 | 35.41 | 0.24 | 0.59 | 0.64 | A−TASSER | 42 | |
| 43 | GS−KudlatyPred | 28.93 | 25.52 | 38.06 | 0.50 | 0.69 | 1.28 | 28.93 | 25.52 | 38.06 | 0.21 | 0.43 | 0.99 | GS−KudlatyPred | 43 |
| 44 | 3D−JIGSAW_V3 | 28.33 | 21.27 | 0.71 | 0.42 | 28.33 | 21.82 | 1.34 | 0.13 | 3D−JIGSAW_V3 | 44 | ||||
| 45 | AMU−Biology | 28.21 | 24.84 | 31.55 | 0.40 | 0.58 | 0.46 | 28.81 | 25.36 | 31.55 | 0.19 | 0.40 | 0.14 | AMU−Biology | 45 |
| 46 | ZicoFullSTP | 27.98 | 21.15 | 34.85 | 0.37 | 0.88 | 39.76 | 29.92 | 44.28 | 1.75 | 1.19 | 1.79 | ZicoFullSTP | 46 | |
| 47 | Kolinski | 27.50 | 23.18 | 30.35 | 0.30 | 0.31 | 0.31 | 29.41 | 25.52 | 32.66 | 0.28 | 0.43 | 0.29 | Kolinski | 47 |
| 48 | SAM−T02−server | 27.14 | 23.81 | 0.26 | 0.41 | 27.74 | 23.81 | 1.03 | 0.04 | 0.13 | SAM−T02−server | 48 | |||
| 49 | circle | 27.14 | 23.53 | 27.80 | 0.26 | 0.36 | 27.26 | 23.53 | 28.20 | 0.08 | circle | 49 | |||
| 50 | FAMSD | 27.14 | 23.53 | 27.80 | 0.26 | 0.36 | 27.14 | 23.53 | 27.80 | 0.08 | FAMSD | 50 | |||
| 51 | Pcons_dot_net | 27.14 | 23.14 | 23.84 | 0.26 | 0.30 | 28.93 | 25.52 | 38.06 | 0.21 | 0.43 | 0.99 | Pcons_dot_net | 51 | |
| 52 | SMEG−CCP | 27.14 | 22.22 | 28.44 | 0.26 | 0.15 | 0.07 | 27.14 | 22.22 | 28.44 | SMEG−CCP | 52 | |||
| 53 | 3DShotMQ | 26.91 | 18.37 | 16.96 | 0.23 | 26.91 | 18.37 | 16.96 | 3DShotMQ | 53 | |||||
| 54 | LOOPP_Server | 26.67 | 23.33 | 27.41 | 0.19 | 0.33 | 27.50 | 23.33 | 27.41 | 0.01 | 0.04 | LOOPP_Server | 54 | ||
| 55 | 3D−JIGSAW_AEP | 26.55 | 24.88 | 1.72 | 0.18 | 0.59 | 26.55 | 24.88 | 1.72 | 0.31 | 3D−JIGSAW_AEP | 55 | |||
| 56 | MidwayFolding | 26.19 | 21.91 | 28.72 | 0.13 | 0.10 | 0.10 | 26.19 | 21.91 | 28.72 | MidwayFolding | 56 | |||
| 57 | RAPTOR | 26.19 | 21.91 | 28.72 | 0.13 | 0.10 | 0.10 | 26.19 | 21.91 | 31.71 | 0.17 | RAPTOR | 57 | ||
| 58 | GS−MetaServer2 | 25.00 | 20.44 | 20.43 | 29.64 | 26.67 | 27.60 | 0.31 | 0.63 | GS−MetaServer2 | 58 | ||||
| 59 | GeneSilicoMetaServer | 25.00 | 20.44 | 20.43 | 29.64 | 26.67 | 27.60 | 0.31 | 0.63 | GeneSilicoMetaServer | 59 | ||||
| 60 | fleil | 25.00 | 19.76 | 22.50 | 25.95 | 20.48 | 23.77 | fleil | 60 | ||||||
| 61 | MUFOLD−Server | 24.29 | 20.48 | 20.10 | 24.41 | 20.48 | 20.14 | MUFOLD−Server | 61 | ||||||
| 62 | Bates_BMM | 23.09 | 19.76 | 31.49 | 0.45 | 23.57 | 21.27 | 31.71 | 0.17 | Bates_BMM | 62 | ||||
| 63 | BioSerf | 22.38 | 18.41 | 33.15 | 0.66 | 22.38 | 18.41 | 33.15 | 0.35 | BioSerf | 63 | ||||
| 64 | FALCON_CONSENSUS | 22.26 | 18.85 | 27.14 | 22.50 | 18.97 | 30.93 | 0.06 | FALCON_CONSENSUS | 64 | |||||
| 65 | FALCON | 22.26 | 18.85 | 30.42 | 0.32 | 22.26 | 18.85 | 30.93 | 0.06 | FALCON | 65 | ||||
| 66 | Handl−Lovell | 21.67 | 13.85 | 29.37 | 0.19 | 21.67 | 15.52 | 32.72 | 0.30 | Handl−Lovell | 66 | ||||
| 67 | Zico | 21.43 | 18.33 | 31.71 | 0.48 | 27.98 | 21.15 | 44.28 | 0.08 | 1.79 | Zico | 67 | |||
| 68 | ZicoFullSTPFullData | 21.43 | 18.33 | 31.71 | 0.48 | 39.76 | 29.92 | 44.28 | 1.75 | 1.19 | 1.79 | ZicoFullSTPFullData | 68 | ||
| 69 | Hao_Kihara | 21.31 | 16.86 | 15.64 | 33.21 | 23.61 | 19.97 | 0.82 | 0.09 | Hao_Kihara | 69 | ||||
| 70 | RBO−Proteus | 21.19 | 19.41 | 32.24 | 0.55 | 23.57 | 19.92 | 32.24 | 0.23 | RBO−Proteus | 70 | ||||
| 71 | mufold | 21.07 | 17.14 | 35.82 | 1.00 | 21.55 | 17.82 | 35.82 | 0.70 | mufold | 71 | ||||
| 72 | fais@hgc | 20.95 | 17.66 | 32.93 | 0.64 | 20.95 | 17.66 | 32.93 | 0.32 | fais@hgc | 72 | ||||
| 73 | BAKER−ROBETTA | 20.59 | 18.33 | 35.98 | 1.02 | 28.93 | 25.52 | 38.06 | 0.21 | 0.43 | 0.99 | BAKER−ROBETTA | 73 | ||
| 74 | FOLDpro | 20.36 | 18.77 | 15.87 | 20.36 | 18.77 | 15.87 | FOLDpro | 74 | ||||||
| 75 | MUProt | 20.24 | 16.23 | 16.38 | 26.19 | 19.21 | 26.43 | MUProt | 75 | ||||||
| 76 | MULTICOM−REFINE | 20.12 | 16.03 | 16.41 | 26.19 | 19.21 | 26.43 | MULTICOM−REFINE | 76 | ||||||
| 77 | MULTICOM−CLUSTER | 20.12 | 15.95 | 16.40 | 21.79 | 18.14 | 23.87 | MULTICOM−CLUSTER | 77 | ||||||
| 78 | Sasaki−Cetin−Sasai | 20.12 | 15.68 | 34.01 | 0.77 | 21.19 | 17.86 | 35.47 | 0.65 | Sasaki−Cetin−Sasai | 78 | ||||
| 79 | 3Dpro | 20.00 | 17.70 | 15.49 | 20.36 | 18.77 | 15.87 | 3Dpro | 79 | ||||||
| 80 | ABIpro | 20.00 | 17.62 | 16.38 | 20.00 | 17.70 | 16.38 | ABIpro | 80 | ||||||
| 81 | SAINT1 | 19.88 | 16.15 | 34.33 | 0.81 | 19.88 | 16.15 | 34.33 | 0.50 | SAINT1 | 81 | ||||
| 82 | POEM | 19.76 | 18.45 | 34.23 | 0.80 | 30.00 | 26.39 | 34.23 | 0.36 | 0.58 | 0.49 | POEM | 82 | ||
| 83 | MULTICOM−RANK | 19.29 | 14.37 | 18.14 | 20.12 | 15.87 | 26.38 | MULTICOM−RANK | 83 | ||||||
| 84 | MULTICOM−CMFR | 19.05 | 15.16 | 18.19 | 30.12 | 22.46 | 24.98 | 0.38 | MULTICOM−CMFR | 84 | |||||
| 85 | Distill | 18.33 | 16.71 | 24.22 | 18.33 | 16.71 | 24.29 | Distill | 85 | ||||||
| 86 | forecast | 17.86 | 15.08 | 27.75 | 18.21 | 15.28 | 27.75 | forecast | 86 | ||||||
| 87 | fais−server | 17.62 | 14.48 | 33.86 | 0.75 | 22.50 | 19.17 | 34.02 | 0.46 | fais−server | 87 | ||||
| 88 | MUFOLD−MD | 16.67 | 14.84 | 32.29 | 0.55 | 20.12 | 17.22 | 33.20 | 0.36 | MUFOLD−MD | 88 | ||||
| 89 | 3DShot2 | 16.55 | 15.32 | 13.06 | 16.55 | 15.32 | 13.06 | 3DShot2 | 89 | ||||||
| 90 | keasar−server | 16.55 | 14.96 | 24.42 | 29.29 | 27.22 | 24.42 | 0.26 | 0.72 | keasar−server | 90 | ||||
| 91 | FEIG | 16.55 | 14.88 | 29.57 | 0.21 | 16.79 | 15.99 | 31.00 | 0.07 | FEIG | 91 | ||||
| 92 | POISE | 16.43 | 14.72 | 25.46 | 16.43 | 14.72 | 25.57 | POISE | 92 | ||||||
| 93 | POEMQA | 16.19 | 13.81 | 19.29 | 30.00 | 26.94 | 29.73 | 0.36 | 0.67 | POEMQA | 93 | ||||
| 94 | PRI−Yang−KiharA | 16.07 | 15.32 | 31.09 | 0.40 | 16.07 | 15.32 | 31.09 | 0.09 | PRI−Yang−KiharA | 94 | ||||
| 95 | PSI | 15.95 | 14.25 | 33.05 | 0.65 | 22.26 | 16.59 | 33.84 | 0.44 | PSI | 95 | ||||
| 96 | TJ_Jiang | 15.71 | 14.01 | 29.62 | 0.22 | 17.14 | 15.48 | 30.83 | 0.05 | TJ_Jiang | 96 | ||||
| 97 | mGenTHREADER | 14.64 | 12.50 | 10.76 | 14.64 | 12.50 | 10.76 | mGenTHREADER | 97 | ||||||
| 98 | Jiang_Zhu | 14.29 | 13.61 | 22.57 | 16.91 | 15.87 | 22.57 | Jiang_Zhu | 98 | ||||||
| 99 | huber−torda−server | 13.93 | 11.98 | 6.00 | 14.17 | 13.69 | 6.00 | huber−torda−server | 99 | ||||||
| 100 | RANDOM | 13.65 | 11.16 | 13.65 | 13.65 | 11.16 | 13.65 | RANDOM | 100 | ||||||
| 101 | PS2−server | 13.57 | 12.62 | 24.64 | 18.57 | 13.97 | 24.64 | PS2−server | 101 | ||||||
| 102 | PS2−manual | 13.57 | 12.62 | 24.64 | 18.57 | 13.97 | 24.64 | PS2−manual | 102 | ||||||
| 103 | MeilerLabRene | 13.45 | 13.06 | 24.93 | 17.74 | 14.80 | 27.61 | MeilerLabRene | 103 | ||||||
| 104 | nFOLD3 | 12.86 | 10.28 | 25.35 | 17.62 | 13.65 | 25.35 | nFOLD3 | 104 | ||||||
| 105 | Softberry | 12.74 | 12.42 | 18.93 | 12.74 | 12.42 | 18.93 | Softberry | 105 | ||||||
| 106 | FUGUE_KM | 12.74 | 11.55 | 12.37 | 14.40 | 12.58 | 19.32 | FUGUE_KM | 106 | ||||||
| 107 | Bilab−UT | 12.62 | 11.47 | 13.31 | 14.52 | 13.06 | 21.35 | Bilab−UT | 107 | ||||||
| 108 | Poing | 12.62 | 11.27 | 16.93 | 13.81 | 12.22 | 20.26 | Poing | 108 | ||||||
| 109 | xianmingpan | 12.50 | 11.39 | 23.07 | 12.50 | 11.39 | 23.07 | xianmingpan | 109 | ||||||
| 110 | DistillSN | 12.38 | 9.92 | 9.85 | 14.52 | 13.10 | 10.03 | DistillSN | 110 | ||||||
| 111 | ACOMPMOD | 12.14 | 10.52 | 21.42 | 13.21 | 11.90 | 21.42 | ACOMPMOD | 111 | ||||||
| 112 | Scheraga | 12.14 | 9.96 | 25.26 | 14.88 | 13.69 | 26.27 | Scheraga | 112 | ||||||
| 113 | SAM−T06−server | 11.79 | 9.37 | 6.89 | 30.00 | 25.56 | 17.54 | 0.36 | 0.43 | SAM−T06−server | 113 | ||||
| 114 | TWPPLAB | 11.67 | 11.03 | 9.19 | 11.67 | 11.03 | 9.19 | TWPPLAB | 114 | ||||||
| 115 | StruPPi | 11.67 | 8.25 | 12.39 | 11.67 | 8.25 | 12.39 | StruPPi | 115 | ||||||
| 116 | DelCLab | 10.95 | 8.89 | 1.44 | 10.95 | 10.36 | 1.53 | DelCLab | 116 | ||||||
| 117 | mariner1 | 10.12 | 8.37 | 13.45 | 11.63 | 14.43 | mariner1 | 117 | |||||||
| 118 | schenk−torda−server | 9.88 | 8.49 | 1.57 | 11.31 | 9.72 | 8.02 | schenk−torda−server | 118 | ||||||
| 119 | Pcons_local | 9.88 | 6.67 | 27.14 | 23.14 | 23.84 | 0.01 | Pcons_local | 119 | ||||||
| 120 | rehtnap | 9.52 | 9.29 | 9.52 | 9.29 | rehtnap | 120 | ||||||||
| 121 | panther_server | 9.40 | 8.53 | 9.40 | 8.53 | panther_server | 121 | ||||||||
| 122 | Abagyan | Abagyan | 122 | ||||||||||||
| 123 | BHAGEERATH | BHAGEERATH | 123 | ||||||||||||
| 124 | CBSU | CBSU | 124 | ||||||||||||
| 125 | FEIG_REFINE | FEIG_REFINE | 125 | ||||||||||||
| 126 | FLOUDAS | FLOUDAS | 126 | ||||||||||||
| 127 | Fiser−M4T | Fiser−M4T | 127 | ||||||||||||
| 128 | FrankensteinLong | FrankensteinLong | 128 | ||||||||||||
| 129 | HCA | HCA | 129 | ||||||||||||
| 130 | JIVE08 | JIVE08 | 130 | ||||||||||||
| 131 | LEE−SERVER | LEE−SERVER | 131 | ||||||||||||
| 132 | Linnolt−UH−CMB | Linnolt−UH−CMB | 132 | ||||||||||||
| 133 | NIM2 | NIM2 | 133 | ||||||||||||
| 134 | Nano_team | Nano_team | 134 | ||||||||||||
| 135 | NirBenTal | NirBenTal | 135 | ||||||||||||
| 136 | OLGAFS | OLGAFS | 136 | ||||||||||||
| 137 | Ozkan−Shell | Ozkan−Shell | 137 | ||||||||||||
| 138 | PHAISTOS | PHAISTOS | 138 | ||||||||||||
| 139 | PZ−UAM | PZ−UAM | 139 | ||||||||||||
| 140 | ProtAnG | ProtAnG | 140 | ||||||||||||
| 141 | ProteinShop | ProteinShop | 141 | ||||||||||||
| 142 | Pushchino | Pushchino | 142 | ||||||||||||
| 143 | RPFM | RPFM | 143 | ||||||||||||
| 144 | SHORTLE | SHORTLE | 144 | ||||||||||||
| 145 | ShakAbInitio | ShakAbInitio | 145 | ||||||||||||
| 146 | TsaiLab | TsaiLab | 146 | ||||||||||||
| 147 | UCDavisGenome | UCDavisGenome | 147 | ||||||||||||
| 148 | Wolfson−FOBIA | Wolfson−FOBIA | 148 | ||||||||||||
| 149 | Wolynes | Wolynes | 149 | ||||||||||||
| 150 | YASARA | YASARA | 150 | ||||||||||||
| 151 | YASARARefine | YASARARefine | 151 | ||||||||||||
| 152 | Zhou−SPARKS | Zhou−SPARKS | 152 | ||||||||||||
| 153 | dill_ucsf | dill_ucsf | 153 | ||||||||||||
| 154 | dill_ucsf_extended | dill_ucsf_extended | 154 | ||||||||||||
| 155 | igor | igor | 155 | ||||||||||||
| 156 | jacobson | jacobson | 156 | ||||||||||||
| 157 | mahmood−torda−server | mahmood−torda−server | 157 | ||||||||||||
| 158 | mti | mti | 158 | ||||||||||||
| 159 | mumssp | mumssp | 159 | ||||||||||||
| 160 | psiphifoldings | psiphifoldings | 160 | ||||||||||||
| 161 | ricardo | ricardo | 161 | ||||||||||||
| 162 | rivilo | rivilo | 162 | ||||||||||||
| 163 | sessions | sessions | 163 | ||||||||||||
| 164 | taylor | taylor | 164 | ||||||||||||
| 165 | test_http_server_01 | test_http_server_01 | 165 | ||||||||||||
| 166 | tripos_08 | tripos_08 | 166 | ||||||||||||