T0465
protein yqbN from Bacillus subtilis
Target sequence:
>T0465 yqbN, Bacillus subtilis, 157 residues
MSENQNEKVYDLSFFMPGQTIDAEEVEVPISKRFVDKEGNVVPFIFKAITTDRIDELEKENTTYKNVKGRGRVKELDSQRFYARIAVETTVYPTFKAKELREAYKTEDPVEVAKRVLSVGGEYANWLNKAIEINGFDDDLEDLEEAAKNLEHHHHHH
Structure:
Determined by: NESG
PDB ID: 3dfd
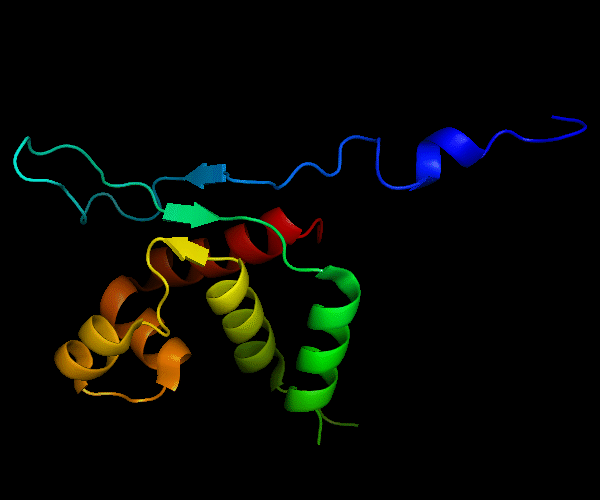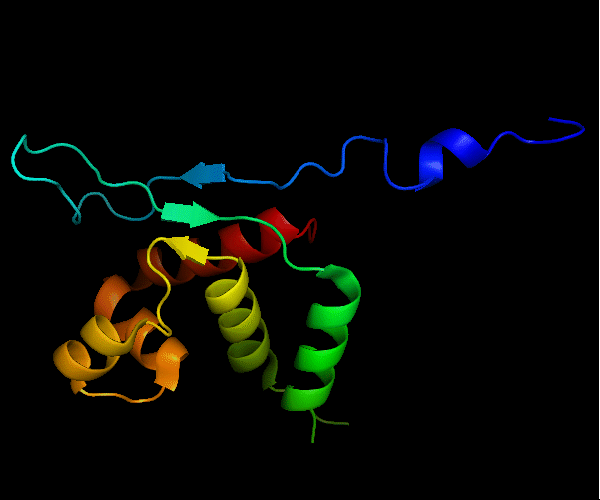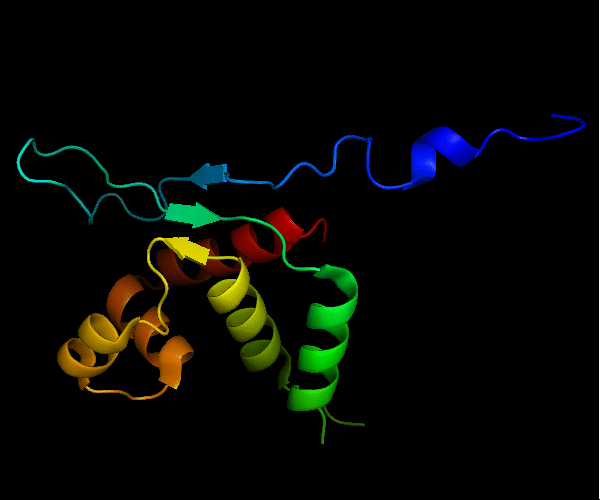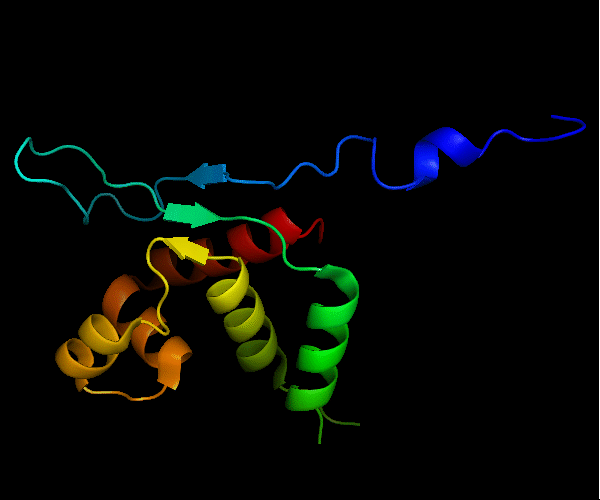
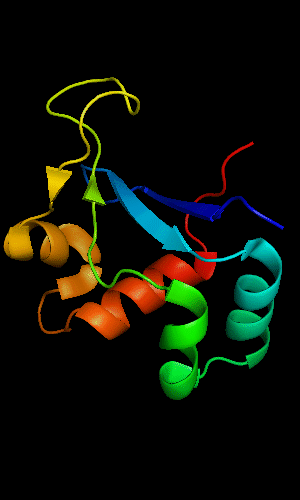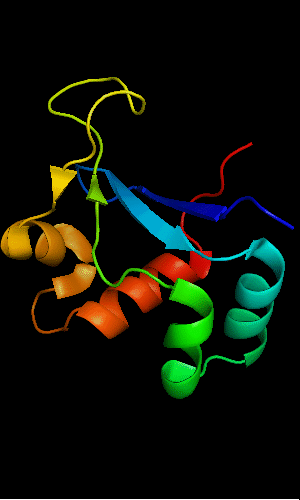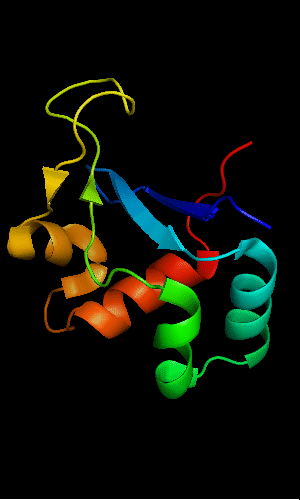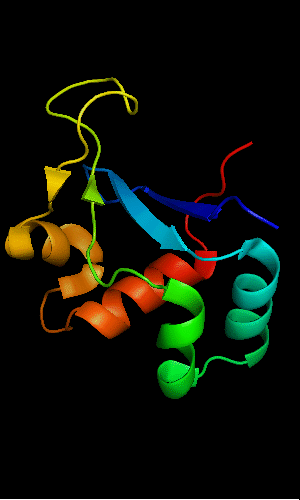
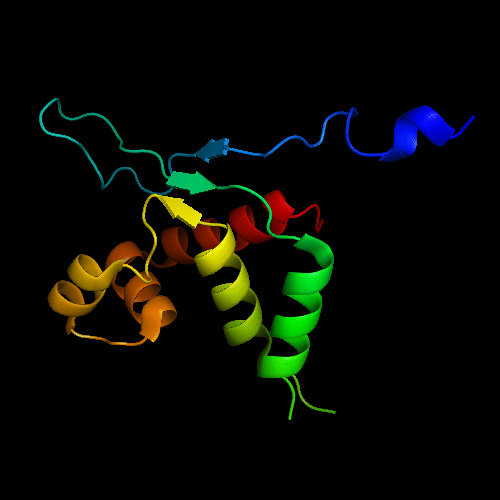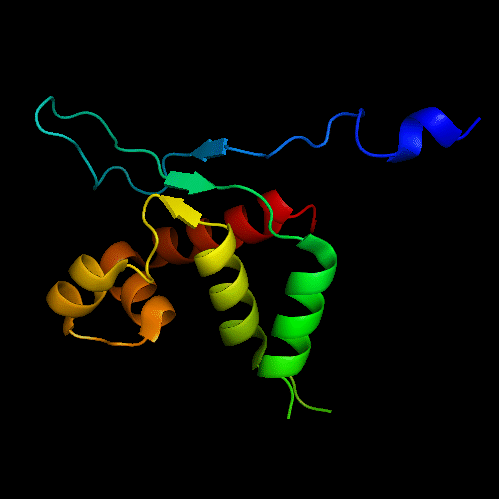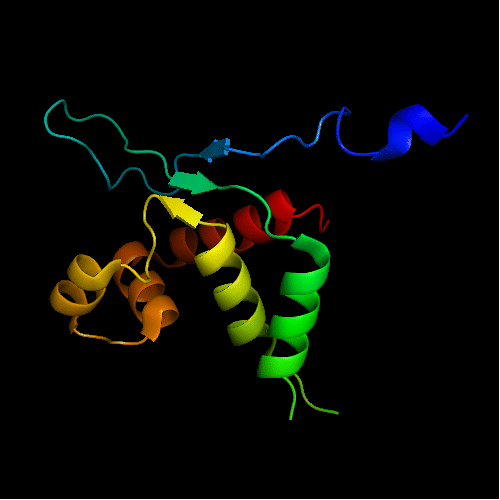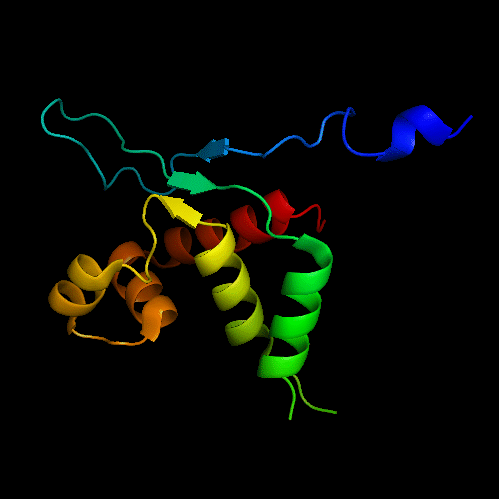
Cartoon diagram of T0465: 3dfd chain A
Domains: PyMOL of domains
Single domain protein with a 14-residue long N-terminal extension 7-EKVYDLSFFMPGQT-20 that forms crystal contacts, but does not interact with its own chain. 6 residues N-terminal to it are disordered.
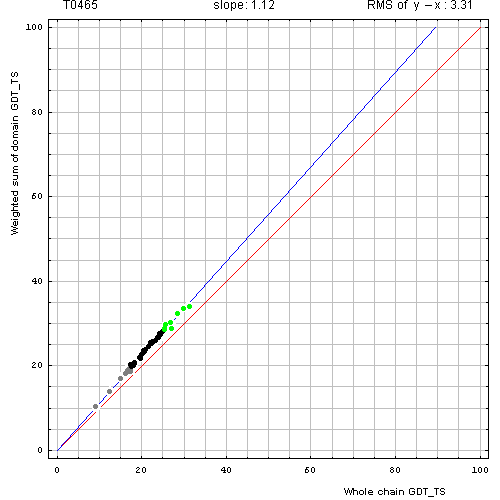
Correlation between weighted by the number of residues sum of GDT-TS scores for domain-based evlatuation (y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
To compute the weighted sum, GDT-TS for each domain was multiplied by the domain length, and this sum was divided by the sum of domain lengths. Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the weighted sum of domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
Mildly distorted FYSH domain. Similar N-terminal β-hairpin (blue-cyan) followed by α-hairpin (cyan-green) and a short β-strand to complete the sheet (lime-yellow). Next β-strand typical of FYSH domain is missing in T0465. Two C-terminal α-helices of FYSH (orange-red) are replaced with three α-helices in T0465.
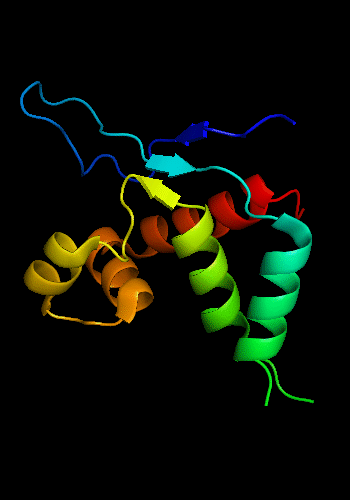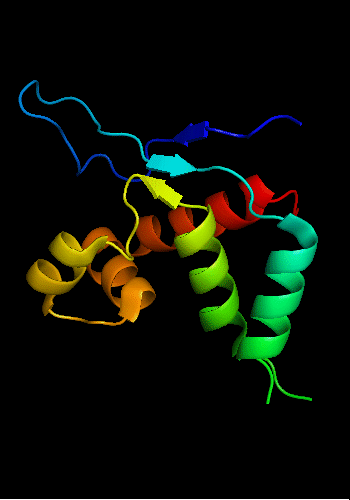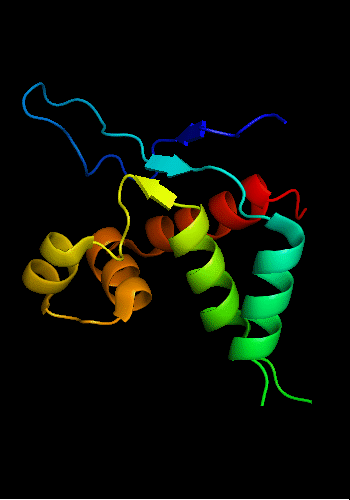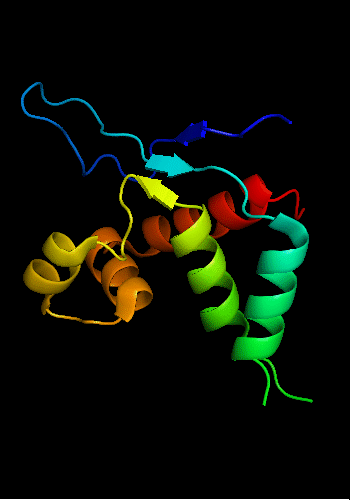
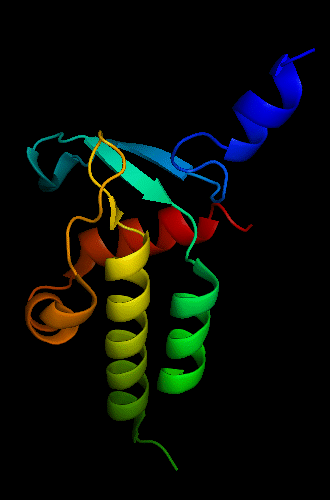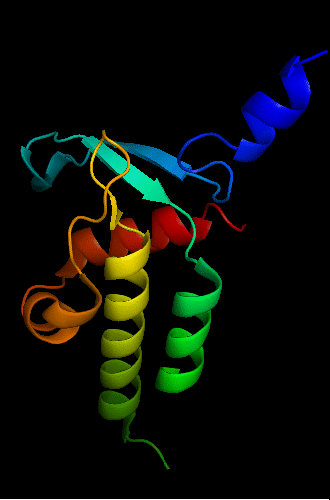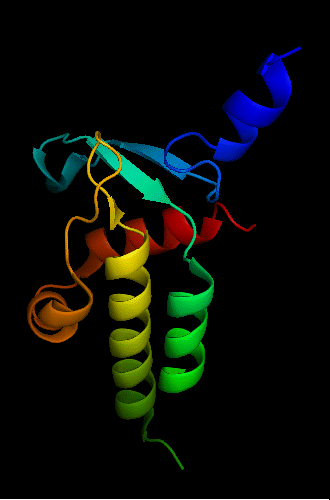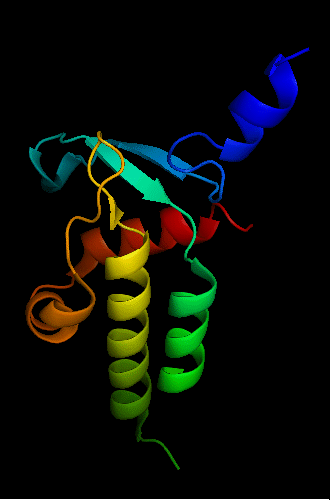
Cartoon diagram of T0465:
3dfd chain A residues 21-136
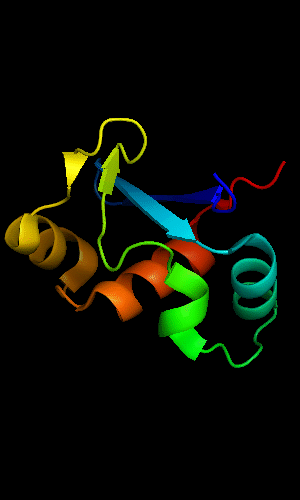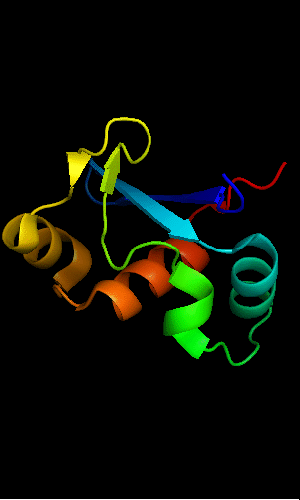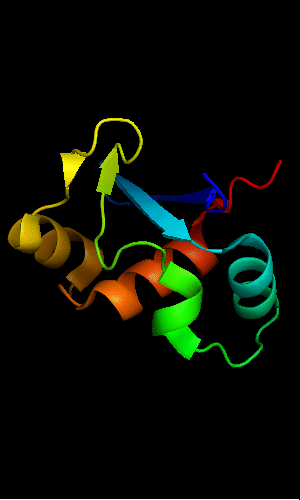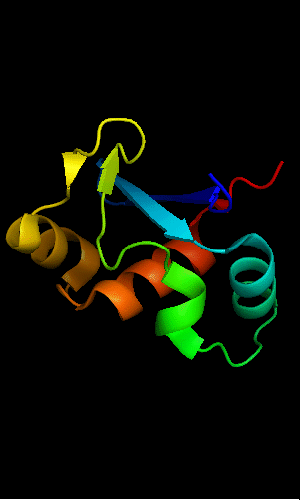
FYSH domain of hypothetical protein AF0491:
1t95 chain A residues 11-94
FYSH domain of hypothetical protein Yhr087W:
1nyn chain A residues 1-93
CASP category:
Free modeling. Fold similarity to FYSH domains not detectable statistically by sequence methods known to us.
Closest templates:
Bacteriophage HK97 tail assembly chaperone 2ob9:
Cartoon diagram of T0465:
3dfd chain A residues 11-137
bacteriophage HK97 tail assembly chaperone:
2ob9 chain A
HHsearch aligns T0465 with FYSH domain protein as the first hit, but with rather low probability (~24%):
>1t95_A Hypothetical protein AF0491; shwachman-bodian-diamond syndrome protein orthologue; 1.90A {Archaeoglobus fulgidus} SCOP: d.235.1.2 Probab=24.32 E-value=14 Score=18.62 Aligned_cols=27 Identities=33% Similarity=0.693 Sum_probs=0.0 Q ss_pred CCHHHHHHCCCCCHHHHHHHHCCCCCHH Q ss_conf 8879998618999899999867996207 Q Mon_Sep_08_03: 96 KAKELREAYKTEDPVEVAKRVLSVGGEY 123 (149) Q Consensus 96 ~d~eLq~sygv~~~~ellk~mL~l~GE~ 123 (149) ++.+|++.||+.+..++++.+| ..||+ T Consensus 65 s~~~L~~~FgT~d~~~I~~~IL-~kGei 91 (240) T 1t95_A 65 SVDELRKIFGTDDVFEIARKII-LEGEV 91 (240) T ss_dssp CHHHHHHHHSCCCHHHHHHHHH-HHSEE T ss_pred CHHHHHHHHCCCCHHHHHHHHH-HCCCC T ss_conf 8899998728998899999999-72885
COMPASS also finds this alignment as 17th hit with E-value of ~20:
Subject = d1t95a2 d.235.1.2 Hypothetical protein AF0491, N-terminal domain length=76 filtered_length=75 Neff=10.627 Smith-Waterman score = 40 Evalue = 1.93e+01 QUERY 96 KAKELREAYKTEDPVEVAKRVL 117 CONSENSUS_1 96 KNKELLEAYKAPDPLELLKKLL 117 ++++L++A+++ D++E++K++L CONSENSUS_2 55 SEEDLKKAFGTTDVDEIIKEIL 76 d1t95a2 55 SVDELRKIFGTDDVFEIARKII 76
Target sequence - PDB file inconsistencies:
6 N-terminal 1-MSENQN-6, 20 C-terminal 138-DDLEDLEEAAKNLEHHHHHH-157 and a loop of 10 residues 64-YKNVKGRGRV-73 are missing from the PDB. Selenomethionine Mse residues are present instead of Met. PDB numbering agrees with the target sequence numbering.
T0465 3dfd.pdb T0465.pdb PyMOL PyMOL of domains
T0465 1 MSENQNEKVYDLSFFMPGQTIDAEEVEVPISKRFVDKEGNVVPFIFKAITTDRIDELEKENTTYKNVKGRGRVKELDSQRFYARIAVETTVYPTFKAKELREAYKTEDPVEVAKRVLSVGGEYANWLNKAIEINGFDDDLEDLEEAAKNLEHHHHHH 157 ~~~~~~|||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~~~~~~~~~~~ 3dfdA 7 ------EKVYDLSFFMPGQTIDAEEVEVPISKRFVDKEGNVVPFIFKAITTDRIDELEKENTT----------KELDSQRFYARIAVETTVYPTFKAKELREAYKTEDPVEVAKRVLSVGGEYANWLNKAIEINGFD-------------------- 137
Residue change log: change 16, MSE to MET;
We suggest to evaluate this target as a single domain spanning whole chain. However, N-terminal segment (target 1-20, pdb 7-20) forms crystal contacts, but does not interact with its own chain. This segment can be removed for domain-based evaluation, resulting this domain definition:
1st domain: target 21-63, 74-137 ; pdb 21-63, 74-137
Sequence classification:
Phage XkdN-like protein family (XkdN) from Pfam. Below is PROMALS3D alignment of homologs detectable by PSI-BLAST. This alignment was not edited manually and contains errors.
Conservation: 9 6 7 5 5 5 5 6 5 7 5 6 89 5 59 6 87 gi_168726643_ref_ZP_02758 1 -M-----------------------NTVEKLLN---MDAGKLKMPS--SIFEL-YCKKLDD-ILEIECNAIDPERFDEIRMNSM---DVDSG---------------NLENINVYELKVNTILESC--PM--------FRNMEVVENLGL-----ATPKDL---LKKLLL-AGEIDNLYEEVNKVNGIDAEGDKK------------------------------------------------------RQKERAKD---IKN------------------------------------------------------------------- 138
gi_168694729_ref_ZP_02727 1 -M-----------------------NTVEKLLN---MDAGKLKMPS--STFEL-YCKKLDD-TLEIECKAIDPERFDEIRMNSM---DVNSG---------------DVENINIYELKVNTILESC--PI--------FSNMELVKKFSA-----ITPKEL---VKKLLL-SGEIDKLYEEVNKVNGIDAEGDKK------------------------------------------------------RQKERAKD---IKN------------------------------------------------------------------- 138gi_90592652_ref_YP_529567 1 -MSENKL----EKEIVDKKEATEVKNIVELLLK---MDAGKIKMPS--MTYKI-FCKKVGI-ELPFECTALEPETFDELQSSGL---KIENG---------------SLKDLDNFKMKTNIILASCKT----------FKDKELLKHFKS-----PTPREL---LRKMLL-AGEINDLYNKICELNGYSESNSE-------------------------------------------------------KDKRIEEK---IKN------------------------------------------------------------------- 156
gi_28211738_ref_NP_782682 1 -M-----------------------NTVEKLLA---LDAGRLEMPN--KEVTLKLGKLGGA-KVTFLCKAVSPEKMAKIQDNLI---EISKKG--------------NIQGTNMGKNKVLTVMEGC-SDT--------FRNKDLLEHFNA-----PTPKEL---INKLLL-AGEIDELYNTINELNGYEQD------------------------------------------------------------EEEEEE---IKNL------------------------------------------------------------------ 136
gi_150018224_ref_YP_00131 1 -M-----------------------NLVEQLLK---IDAGKIEVPS--KEVKL-KLAKLGNMEITFKCNAISMERYNEIQERVL---QVDKKG--------------NIQGFATAQAKIETVLAGV--PE--------LRSEELMKHFKA-----PTPKEL---MNKIFL-PGEVDILADTVTEISGVEST-------------------------------------------------------------NQKED---IKNS------------------------------------------------------------------ 134
gi_126698965_ref_YP_00108 1 -MSEN--------------GLLKNINIVDLLLN---ADTENLERPSTIVELKR-LSTIFGQ-EFKVMCRALTISKDEEIQNTCL---KIDEN---------------MKTDIDLPEMQMLTIIEGVCDLDGK----LLFKNKELMDKFKA-----PTPKEL---ARKLLL-PGEITNLYRILQDVMGYGKN-------------------------------------------------------------AVIEE---VKN------------------------------------------------------------------- 148
gi_145955100_ref_ZP_01804 1 -MNEN--------------GLSKNINIVDLLLN---ADTENLERPSTIVELKR-LSTIFGQ-EFKVMCRALTISKDEEIQNTCL---KIDEN---------------MKTDIDLPEMQMLTIIEGVCDLDGK----LLFKNKELMDKFKA-----PTPKEL---ARKLLL-PGEITNLYRILQDVMGYGKN-------------------------------------------------------------AVIEE---VKN------------------------------------------------------------------- 148gi_168070691_ref_XP_00178 1 ---------------------------------------------------------MKRL-GVDFQIQALDGKTIHKIQEQCT---HYTGKGVK------------REKVLDEEQFGALVIQRACLIPD--------WSARELIDKYG-------SPTEA---ILGVLL-AGEIAKLSSEILEISARGRGEDTAGRSKGCPQENTVGPLTGIYGKRGGGDDWLRLRTTAEAAFRFRLQTTTISLEQFNGIAQMSEEMWKAGDLSAQ--------------------------------------------------------------- 175
gi_150390328_ref_YP_00132 1 -MLDEKNTILEEELTEEEMIEMSEDDIINKLL-------EPSDAP----ERTY-AIERL---GIQVTLKGLSEREIQRIRKECT---VERKHRGN------------RTKELNEEEFNAALIESATIKPN--------WSDKRLLSSLSL-----SSGREV---IKRRLL-AGEMVALGDKVMELSGFDN---------------------------------------------------------------ELEE---IKN------------------------------------------------------------------- 149
gi_148380291_ref_YP_00125 1 MSNEK--------IKDEEILNMTEEDIINRLM-------EPDEVP----EATY-FIERL---RIPVTLKGLSEKEINKIKRQCT---YTRKERGK------------RIKELDDEEFNAALIEAATVTPN--------WNDERLLDALKA-----SDGKQV---IRKKFL-AGETSAMGDKVLELSGFDN---------------------------------------------------------------ELEE---IEDIKN---------------------------------------------------------------- 145gi_187932802_ref_YP_00188 1 -MN-----------TENNVLEMQEEDIIKKLL-------GEVEVP----KAVV-VIERI---GIPVELKGLNTVEIKKIREKCT---SKRKIKGV------------AEEKLNGEEYDAALIIGATTNFD--------WNNPKLLEKYSL-----SDGKQF---ILRKLL-AGERTNLVNKVLDLSGYNDEL------------------------------------------------------------SDAED---IKNLSGEEE------------------------------------------------------------- 147
gi_153955865_ref_YP_00139 1 -MKKM---------TDEQILSMKEEDILAKLM-------GTYEVP----TATV-FIERM---GIPLTLKGLSEKEISKIRKECT---YNRKSRGGK-----------SEDKLDGNEFDAGLIVAATTNFN--------WNDSKLLDSAKA-----SDGKQF---IRKKLL-AGEISALTDKILELSGFNNEL------------------------------------------------------------EEMED---IKNS------------------------------------------------------------------ 145
gi_150016525_ref_YP_00130 1 -MKKN---------EKDEVLAMKEEDILAKLL-------ETHDVP----TANI-QVPRL---GIAIDLKGLTEKEISGIREECT---TRRK-INGK-----------IETKLNNADFDAALIIGATTNFN--------WNNPKLIEPLKL-----SDGKAY---IRKKLL-AGEISFLANKVLELSGFNDEL------------------------------------------------------------EEKED---IKN------------------------------------------------------------------- 143gi_153954477_ref_YP_00139 1 -MD------------IEKMQNMSEDDVIDALL-------GEIEVP----TRTV-VIQRL---GIPIKLKALTGKQISKIRKDNT---HSEKVKGSKL----------EKDVFDDENFNAEIIEKATVSPN--------WNNEKLKAALNV-----SNGKEV---IKRRLL-AGEMDNLIEQVFDLSGYNDEA------------------------------------------------------------EDIEE---IKNSLEPDTDSI---------------------------------------------------------- 151
gi_153954568_ref_YP_00139 1 -MD------------IEKMQNMSEDDIIDALL-------GEIEVP----TRTV-VIQRL---GIPIKLKALTGKQISKIRKDNT---HSEKVKGSKL----------EKDVFDDENFNADIIEKATVSPN--------WNNEKLKAALSV-----SNGKEV---IKRRLL-AGEMDDLINKIFDLSGYNDEA------------------------------------------------------------EDIED---IKNSSGLDTNFI---------------------------------------------------------- 151gi_192811458_ref_ZP_03040 1 -MS------------LHE--NMSEEQILDSLF-------EAAERL---PEETV-RIKRL---DMQMVLQGLTSSKVDSIRERCT---VRRTVKGT------------VDEKVDTETFNALLISEATSSLSVKGLTLSGWGDPRITSRLKL-----SGGEQA---VRRMLL-AGELDAVGDKVLELSGFGV---------------------------------------------------------------EIAD---LKN------------------------------------------------------------------- 144
gi_187604630_ref_ZP_02990 1 -MA-----------VKNK-----FVSLEDVLG---KNAEELTAVT----LGEI-DAKKL---GGKLPVASFDNKEFSQIKKDCT---TYVKSGKNGR----------MLPEIDEDKLMVEVIVAAVHNDTRS---DFTFRNPQLLEKLGV-----ISAAEA---AGILLS-PGEIQRGAMEVQEISGFNTGAE----------------------------------------------------------EERAEE---VKNS------------------------------------------------------------------ 150
gi_182418566_ref_ZP_02626 1 -MQ---------------------LTIENLLES-KGIIEGETGVK----TTNL-GIKRL---GGEITIQSLDPKKLEQILKEAN---SGKS----------------------TLELNKKIIYMSVIDPN--------LKDNELLKGYGC----KNNPYEI---VEKIFT-ITEIGIIADKIAKLNGLDDVKN---------------------------------------------------------LEDFVVE---IKN------------------------------------------------------------------- 131
gi_150390186_ref_YP_00132 1 -MAKK--------PVIKK------LTLNDLMQQ--REKYEVKQDT----KEEV-LLRRGDD-QVTITIKKPTRS----LCLECI---TLSQ------------------DENRQEQADINMVYNIVVEPN--------LKDTNLHKAYGC-----IEPIEI---VEKIFE-TGEIAQISGYGMELAGYGNE------------------------------------------------------------MKAIKD---IKN------------------------------------------------------------------- 135
gi_109648029_ref_ZP_01371 1 -M-----------------------DTLELLLKGERPNMP-EKEI----KLKR-LSKACGG-DIIFRLRALSFNRVAEIKNSHA-----------------------------GGDMEVHILLAGVISPD--------LKSEDLKQKYNA-----VTPLEM---IKTMLL-PGEIEDISREIEKLSGYRVT--------------------------------------------------------------TVEE---VKKK------------------------------------------------------------------ 122
gi_90592622_ref_YP_529882 1 -MAE---------------------SVEDFLFE--NVGSP---------VEEK-EIKLERF-KSPFKIKSLTADEVSELRKQAT---KRVLNRK-THK---------YEQDTDENQFQDLVVAEAVVSPN--------LNNEELQTSWGC----IAKPEKV---LKKMLK-VGEYTELSQAIMDLSGLNDDDS---------------------------------------------------------SEDLVEE---AKN------------------------------------------------------------------- 139
gi_163932194_ref_YP_00164 1 -MAE---------------------SVEDFLFE--NVGSP---------VEEK-EVKLERF-KSPFKIKSLTADEVSDLRKQAT---KRVLNRK-THK---------YEQETDENQFQDLVVAEAVVSPN--------LNNEKLQTSWGC----IAKPEEV---LKKMLK-VGEYTELSQAIMDLSGLNDDDS---------------------------------------------------------SEDLVEE---AKN------------------------------------------------------------------- 139gi_116629278_ref_YP_81445 1 -MAE---------------------SVEDFLFE--NVGSP---------VEEK-EVKLERF-KSPFKIKSLTADEVSHIRKQAT---KRVLNRK-THR---------YEQETDENQFQGLVVAEAVVSPN--------LNNEKLQTSWGC----IAKPEEV---LKKMLK-VGEYTELSQAIMDLSGLNDDDS---------------------------------------------------------SEDLVEE---AKN------------------------------------------------------------------- 139gi_39653729_ref_NP_945294 1 -M-----------------------AISDFLLE--NVQQE----------ETK-EVHLKRF-KSPFVIRSIDESLNDTLKKRAT---IKKKNRQGV-----------AIPEFNNDKYIDSLMSACVVTPD--------LKDAQLQESYRT----VGDEAAT---LKAMLK-IGEYATLMQEIQSLNGFDEDI-----------------------------------------------------------NDLVEE---SKKRLEDGDAELSYAYYCLHQFNWTPSFLDSLSKREKALIFAFIDIRVEAEEKEHKEMERKSKGRRRR-- 198
gi_169185014_ref_ZP_02845 1 -M-----------------------NLQEFLNN-HPVDNLTEEII----VSPR-FKDADGL-PLKFTIKAMTSQEFEEIRKSAT---EIRKG---------------RKVEFDAQKFNLRAVINHTIVPD--------FKDAASIQKLGC-----RTADEY---VQKVLL-AGEMSTIVSKIQELSGFDVGM-----------------------------------------------------------NELIDE---AKN------------------------------------------------------------------- 135
gi_153941040_ref_YP_00139 1 -MN----------------------NFEDFLMD-SFEEVEEIER----------EITIGGK-KKKMKFKPISADKGDELRKKCK---KVKLVKGQ------------KMIETDQDKFVSNQIIETTIYPD--------LKNSELQKAWGV-----IGAEELLSAMKSRMT-DGEYADWSSTVIEINGYDKGM-----------------------------------------------------------QELVEE---AKN------------------------------------------------------------------- 137
gi_168179424_ref_ZP_02614 1 -MN----------------------NFEDFLMD-SFEEVEEIER----------EITIGGK-KKKMKFKPISADKGDELRKKCK---KVKLVKGQ------------KMIETDQDKFVSNQIIETTIHPD--------LKNSELQKAWGV-----MGAEELLNAMKSRMT-DGEYADWSSTVIEINGYDKGI-----------------------------------------------------------QELVEE---AKN------------------------------------------------------------------- 137gi_148379694_ref_YP_00125 1 -MN----------------------NFEDFLMD-SFEEVEEIER----------EITIGGK-KKKLKFKPISADKGDELRKKCK---KITIVKGQ------------KMSETDQDKFIANQIIETTVYPD--------LKNAELQKAWGV-----MGAEQLLKAMKSKMS-DGEYMGWGSVVSEINGYDKGI-----------------------------------------------------------QELVEE---AKN------------------------------------------------------------------- 137gi_168178739_ref_ZP_02613 1 -MN----------------------NFEDFLMD-SFEEVEEIER----------EITIGGK-KKKMKFKPISADKGDELRKKCK---KITIVKGQ------------KMSETDQDKFIANQIIETTVYPD--------LKNAELQKAWGV-----MGAEQLLKAMKSKMS-DGEYMEWGSVVSEINGYDKGI-----------------------------------------------------------QELVEE---GKN------------------------------------------------------------------- 137gi_188587578_ref_YP_00192 1 -MN----------------------NFEDFLMD-SFEDTQEIER----------EVTIGGK-KKLMRFRPISAEMGDMIRKRNR---KTKLIKGQ------------RIMETDQDKYVSDLIIETTTCPD--------LKNSELQASWGV-----LGAEELLSAMKSKMR-DGEFSDWSSIVGEVNGYDKSV-----------------------------------------------------------NDLIEE---AKN------------------------------------------------------------------- 137gi_168185999_ref_ZP_02620 1 -MS----------------------QFEDFLID-SFEDVEEVEK----------ELEIGGR-KRKMRFKAIGATRADEIRRDCR---NVKFIKSQ------------KIVETDQDKYLEKVIVETTVYPD--------LKNAELQSNWGV-----MGAVELLQAMKSKMT-DGEYSSWSNVVSEINGYDKEL-----------------------------------------------------------QELIEE---AKN------------------------------------------------------------------- 137gi_89896701_ref_YP_520188 1 -MS----------------------DLQDFLMD-NFEEAEPIER----------RVSLGGK-EKIMKFKPISAATGDEIRRSCR---KTTFHKGQ------------RLVETDQDAFVAKLIIETTTVPD--------FKSQELQQSWGVL--GAENLLKA---MKTKMK-DGEYATFSNIVSEVNGYDKSM-----------------------------------------------------------NDLVEE---AKN------------------------------------------------------------------- 137
gi_169827347_ref_YP_00169 1 -MS----------------------NLTAFFAH-NKKQNENIKHA----ISKK-FVDEQGN-PIEWEFAPISPERDEELKSEST---KRSMIMQGKRKG-------QYNTDFDHFKYQRLLTIESIVYPN--------LNDKELQDSYGV-----MGADAL---LGKMLT-IGEIADASAVAQEVNGYQAEL-----------------------------------------------------------EDMVEE---IKN------------------------------------------------------------------- 144
gi_126650061_ref_ZP_01722 1 -MS----------------------NLTAFFAH-NKKQNENIKHA----ISKK-FVDEQGH-PIEWEFAPISPERDEELKSEST---KRSMIMQGKRKG-------QYNTDFDHFKYQRLLTVESIVYPN--------LNDKELQDSYNV-----MGADAL---LGKMLT-IGEIADASAVAQEVNGYQAEL-----------------------------------------------------------EDMVEE---IKN------------------------------------------------------------------- 144gi_126698538_ref_YP_00108 1 -MS----------------------NLSAFLSQ-NAIKVDNVKYV----ASNR-FLDKEGK-PVEWELKVLSSEEDEALRRKCT---KRVKVIGNNGKHT-----GQYTSEIDYNSYVAELCVASTVFPD--------LKDAELQNSYGV-----MGEAQL---LKTMLT-AGEYVNYTVKVNEVNGFDTSF-----------------------------------------------------------EDKVEE---AKN------------------------------------------------------------------- 146
gi_168708891_ref_ZP_02741 1 -MG----------------------DLNAFLSQ-NAIKVENRKYV----ASNR-FINEEGK-AIEWEIRALSSEEDAAIRKNCP---KREPILNKKGKHT-----GQYNTVTDYNKYYEELSIACTVFPD--------LNDSMLQDSYRV-----MGANQL---LKAMLT-PGEYTEYVQEVLDINGFDNSF-----------------------------------------------------------EDKVEE---AKN------------------------------------------------------------------- 146gi_134287351_ref_YP_00111 1 -MG----------------------DLNAFLSQ-NAIKVENRKYV----ASER-FIGEDGK-AIEWELKAIDSDRDRQLRKDST---IRVPVLNKKGKAT-----GQYTSETDFNTYTLKLCVETVVFPD--------LHDAELQNSYGV-----MGAEEL---LTTMLT-PGEYTDLSSEVGEVNGFDRTF-----------------------------------------------------------EDKVEE---AKN------------------------------------------------------------------- 146gi_168714500_ref_ZP_02746 1 -MS----------------------NLSAFLSQ-NAIKVDNVKYI----ASDR-FLDEEGK-PVEWELKVLSSEEDEVLRRNCT---KRVKVIGNNGKPT-----GQFTSEIDYNSYVAELCVASTVFPD--------LKDAELQNSYGV-----MGEAQL---LKTMLT-AGEYVNYTVKVNEVNGFDTTF-----------------------------------------------------------EDKVEE---AKN------------------------------------------------------------------- 146gi_167770421_ref_ZP_02442 1 -MKMA-------------------GGLSAFLAQ-NAVKPENIRFA----ASRR-FVDEDGK-PMLWELRPLMSEEDEALRRRFT---RMVQVPGKSSQ---------FRQEFDSNGYLAAMAAGCTVFPN--------LNDKGLQDSYGV-----MGAEAL---LKAMLT-SGESANYFQKAQEVCGFTPI------------------------------------------------------------EELTEQ---AKN------------------------------------------------------------------- 144
gi_125974972_ref_YP_00103 1 -MSL--------------------GSLEAFLNP---VKVENKKVI----VSNR-FKDKDGN-LVPFEIRPITQEENKMLIKKYT---KRDKK---------------GQEYFDRAEYISELTASAVVFPD--------LTNAELQKAYGV-----LGASAL---LQKMLY-VGEYAELAQAVQELSGLDTDI-----------------------------------------------------------NEDIEE---VKNA------------------------------------------------------------------ 137
gi_167462289_ref_ZP_02327 1 -MS---------------------GGLQAFFAQ-EAGEAITEKVV----VSDR-FKDKDGK-PVPWEIRSMTEAENEEIRKSAT---RKVKSK--NGM---------YTTETNNELYLAKLAVASVVFPD--------LKNVELQQSYGT-----LGAENL---LRRMLL-PGEYANLIEKVQEINGFDKDI-----------------------------------------------------------EDLKEE---IKN------------------------------------------------------------------- 141
gi_167463522_ref_ZP_02328 1 -MS----------------------DLSLFYAQ-NAEAGIMEEFA----VSNR-FKTKDGK-PVLWKIRCMTEAENEEHRKAAT---KKVKAK--NGT---------YLSETNSDLYLAKLAVGSVVYPD--------LKDAELQKSYGT-----LGAENL---LRVMLL-PGEYAALIEKVQQINGFDKDL-----------------------------------------------------------NELKEE---VKN------------------------------------------------------------------- 140gi_192811441_ref_ZP_03040 1 -MS----------------------DFSMFFAG-QSSAEITEEFV----VSVR-FKNAEGS-PVPWKLRSITEEENQECRKAAT---RKVKGK--NGV---------FTPEIDPNDYMAKLMVSSVIYPD--------LKNSELQKSYGV-----LGAESL---LRKMLL-PGEFAALGERVQALNGFDRDM-----------------------------------------------------------NDLVDD---VKN------------------------------------------------------------------- 140
gi_160878945_ref_YP_00155 1 -M-----------------------SLQLFMKK-NKKVKENVFYA----PTKS-LLDENGL-PLNWEFRHVSTKEDEDIRESCT---MDVQITGKPGA---------YRKKIDTNAYIAKLVAASCVVPN--------LNNAELQDSYGV-----KKPEDL---LKELVDDPGEYQDLFVFIQKYNGFDTSM-----------------------------------------------------------EEEVEE---AKN------------------------------------------------------------------- 142
gi_160881084_ref_YP_00156 1 -MS----------------------NFNRFMKQ-NKVQKENTTYP----ATKS-LTDEKGN-PLLWSIKPLTTRENDAIREDCT---MDVPVTGKPNM---------FRPKVNTSKYIAKMICAAVIEPN--------LNDKELQDSYGV-----MTPEEL---LKEMVDDPGEYGDFANFVQQFNGFTTTM-----------------------------------------------------------DDKVEE---AKN------------------------------------------------------------------- 143
gi_153810680_ref_ZP_01963 1 -MGGDNPP---YNFEKMEEKRMG-IDMKAFMKP-ELKDRGTMEFP----GIER-YKDAKGN-VIPFIIKRLSMKEIKEIRNNYKTKEVYRDKRNGDRPIIGNNGQVAVINDYDGDSAGLEIMVEAFVQPK--------LDDPELMEYYGV-----HDRLDM---PNIIFPDKDDFKYADDCLMEACGLASKKND--------------------------------------------------------KETVEEI---KKMMSGEGDDENSFDWMWAHVLWQKKGLRMEEYAMMERNVRLAYIASEELAIKAPLNADERLARGFLKKK 243
gi_16800349_ref_NP_470617 1 -MVADREI---EKVEESN--EELVYDITAFLPG-KHEEKVKLERA----VSNR-FKDKNGN-VIKFVFENVTSERLEELRKDCT---KRIPKGR-GRQ---------PEEKFDSDKFQALLALESTVYPK--------FSSKELLNAYKS-----PSALEV---AKKVLSVPGDYMNWISAASEANGFDDEIDD--------------------------------------------------------EAESIEL---AKN------------------------------------------------------------------- 162
gi_16078332_ref_NP_389149 1 -MSEK---------------NENVYDLSFFMPG-KTIEAEEIKVP----ISKR-FVDKKGN-VIPFILKPITTERIDELEKENT---TFKNVKSRG-----------RVKDLDSQRFYARIAIESTIYPD--------FRSKDLREAYKT-----ADPVEV---AKRVLSVGGEYANWLNKAIEINGFEDDL-----------------------------------------------------------EDLEEE---GKKLVKDGDKEAVYLYYAMHELHYSPSQLLELYEAPKHFKALLFGLIGYKLDLLEKESRRGGN------- 208
gi_16079657_ref_NP_390481 1 -MS------------ENQ--NEKVYDLSFFMPG-QTIDAEEVEVP----ISKR-FVDKEGN-VVPFIFKAITTDRIDELEKENT---TYKNVKGRG-----------RVKELDSQRFYARIAVETTVYPT--------FKAKELREAYKT-----EDPVEV---AKRVLSVGGEYANWLNKAIEINGFDDDL-----------------------------------------------------------EDLEEA---AKN------------------------------------------------------------------- 149T0465_yqbN,_Bacillus_subt 1 -MS------------ENQ--NEKVYDLSFFMPG-QTIDAEEVEVP----ISKR-FVDKEGN-VVPFIFKAITTDRIDELEKENT---TYKNVKGRG-----------RVKELDSQRFYARIAVETTVYPT--------FKAKELREAYKT-----EDPVEV---AKRVLSVGGEYANWLNKAIEINGFDDDL-----------------------------------------------------------EDLEEA---AKNLEHHHHHH----------------------------------------------------------- 157
gi_109648029_ref_ZP_01371 1 -M-----------------------DTLELLLKGERPNMP-EKEI----KLKR-LSKACGG-DIIFRLRALSFNRVAEIKNSHA-----------------------------GGDMEVHILLAGVISPD--------LKSEDLKQKYNA-----VTPLEM---IKTMLL-PGEIEDISREIEKLSGYRVT--------------------------------------------------------------TVEE---VKKK------------------------------------------------------------------ 122
gi_167745915_ref_ZP_02418 1 -MGNAAK-----AEEMVKVTKENELDLVQGLLE--AADFRNDDDL----QTEI-QIKRNDKVFFKFKVRPLSEEELVKARRNAV---VKMPNPSNPKLP-------PIEKDLRVEEFTAWKIYYATVDKEE------VWDNPSVMKGLEAKGFPVLRGIDV---INTVLR-AGEKDAIDDQIDKISGFYDSE-----------------------------------------------------------ISEEEY---AKN------------------------------------------------------------------- 168
gi_167767968_ref_ZP_02440 1 -MAEKK-------ETNVTVTEDNEMDLITGLLK--AAEYKTEV------QQPL-NITRNGQTLFKFNVRPLSFDEIAQCRKKAT---TYMANPGGASLP-------LVEKEVSTADYMAWKIYTATVATDGK----KFWDNSALKEGLKKAGHMVMTQNEI---IKEVLT-AGELEAVSDAIDNLSGGGVS--------------------------------------------------------------VVDY---AKN------------------------------------------------------------------- 163
Consensus_aa: .Ms......................sh..bh.p....p..p.c........p....p..s.....h.hpsls.pc.pplppphh.........................pphs..ph...hhh.thh.ss........hps.pLbp.h.h......ss.ph...hpp.h..sGEhs.h.p.l.clsGhs.p.............................................................p..--...hKN...................................................................Consensus_ss: hhhhhhh hhhh hh eeeeeee hhhhhhhhhhh ee hhhhhhhhhhhhh hhhhhh hhhh hhhhh hhhhhhhhhhhhh h hhhhhh h
Comments:
Very interesting structure and a nice CASP target!
Server predictions:
T0465:pdb 7-137:seq 7-137:FM; alignment
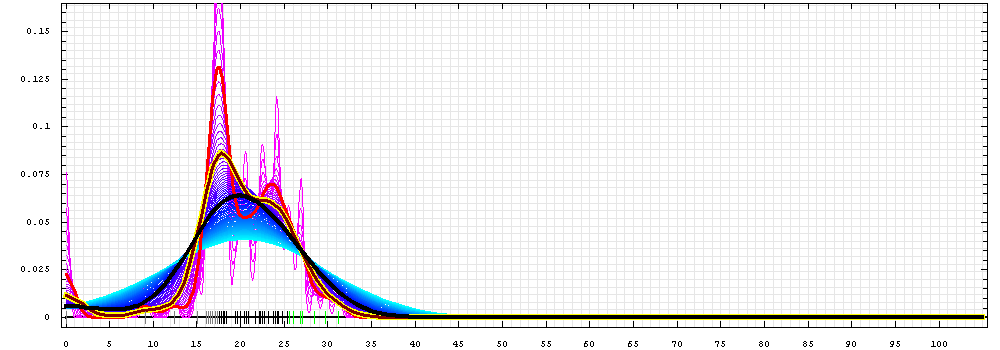
First models for T0465:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
465_1:pdb 21-137:seq 21-137:FR; alignment
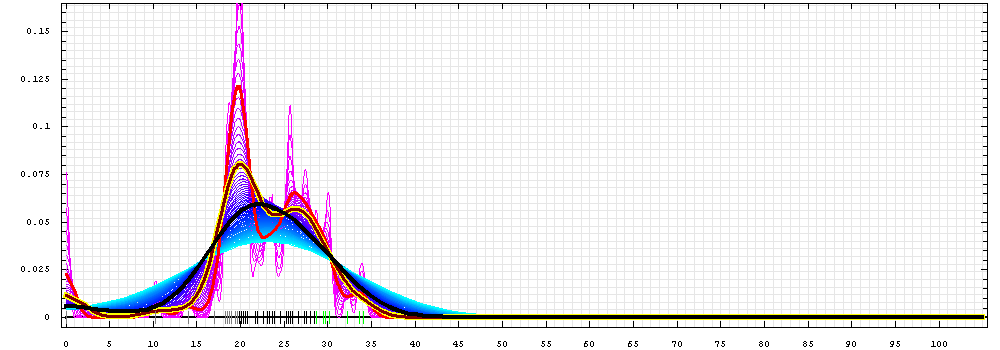
First models for T0465_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0465 | 465_1 | T0465 | 465_1 | T0465 | 465_1 | T0465 | 465_1 | ||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||
| 1 | ABIpro | 31.20 | 23.76 | 44.73 | 34.11 | 25.70 | 46.21 | 2.11 | 1.62 | 1.18 | 1.91 | 1.34 | 1.16 | 31.20 | 23.97 | 45.55 | 34.11 | 25.70 | 47.20 | 1.66 | 1.30 | 1.06 | 1.46 | 0.91 | 1.04 | ABIpro | 1 |
| 2 | FALCON | 31.20 | 23.76 | 44.73 | 34.11 | 25.70 | 46.21 | 2.11 | 1.62 | 1.18 | 1.91 | 1.34 | 1.16 | 31.20 | 24.73 | 46.74 | 34.11 | 27.96 | 48.40 | 1.66 | 1.57 | 1.19 | 1.46 | 1.64 | 1.18 | FALCON | 2 |
| 3 | SAM−T08−human | 30.58 | 22.93 | 46.73 | 33.88 | 25.31 | 48.23 | 1.96 | 1.35 | 1.39 | 1.86 | 1.23 | 1.39 | 30.58 | 22.93 | 46.73 | 33.88 | 25.31 | 48.23 | 1.50 | 0.93 | 1.18 | 1.41 | 0.79 | 1.16 | SAM−T08−human | 3 |
| 4 | Zhang | 30.37 | 24.59 | 47.83 | 34.11 | 28.35 | 48.61 | 1.91 | 1.89 | 1.51 | 1.91 | 2.11 | 1.43 | 30.37 | 24.59 | 48.76 | 34.11 | 28.35 | 51.34 | 1.45 | 1.52 | 1.41 | 1.46 | 1.76 | 1.52 | Zhang | 4 |
| 5 | fams−ace2 | 30.16 | 23.97 | 46.39 | 34.11 | 27.10 | 47.48 | 1.86 | 1.68 | 1.36 | 1.91 | 1.75 | 1.31 | 31.20 | 25.00 | 46.69 | 34.35 | 27.57 | 48.26 | 1.66 | 1.66 | 1.18 | 1.51 | 1.51 | 1.16 | fams−ace2 | 5 |
| 6 | mufold | 29.96 | 23.28 | 45.18 | 32.94 | 25.47 | 46.66 | 1.81 | 1.46 | 1.23 | 1.66 | 1.27 | 1.21 | 30.99 | 23.28 | 45.18 | 34.81 | 26.25 | 46.66 | 1.60 | 1.05 | 1.01 | 1.61 | 1.09 | 0.98 | mufold | 6 |
| 7 | Jones−UCL | 29.75 | 24.93 | 47.70 | 32.94 | 27.49 | 49.42 | 1.76 | 2.00 | 1.49 | 1.66 | 1.86 | 1.52 | 29.75 | 25.55 | 47.70 | 32.94 | 28.43 | 49.42 | 1.30 | 1.86 | 1.29 | 1.20 | 1.79 | 1.29 | Jones−UCL | 7 |
| 8 | Zhang−Server | 29.75 | 23.42 | 45.21 | 33.65 | 26.48 | 46.79 | 1.76 | 1.51 | 1.23 | 1.81 | 1.57 | 1.23 | 29.75 | 24.38 | 45.21 | 33.65 | 27.34 | 47.92 | 1.30 | 1.44 | 1.02 | 1.36 | 1.44 | 1.12 | Zhang−Server | 8 |
| 9 | keasar | 29.13 | 22.04 | 44.51 | 32.24 | 24.69 | 46.33 | 1.61 | 1.06 | 1.16 | 1.50 | 1.05 | 1.18 | 29.34 | 22.31 | 45.02 | 32.71 | 25.86 | 46.91 | 1.20 | 0.71 | 1.00 | 1.15 | 0.97 | 1.00 | keasar | 9 |
| 10 | Zico | 28.93 | 23.42 | 47.66 | 32.48 | 26.25 | 49.59 | 1.56 | 1.51 | 1.49 | 1.56 | 1.50 | 1.54 | 30.99 | 23.42 | 47.66 | 35.05 | 26.25 | 49.59 | 1.60 | 1.10 | 1.29 | 1.66 | 1.09 | 1.31 | Zico | 10 |
| 11 | ZicoFullSTP | 28.93 | 23.42 | 47.66 | 32.48 | 26.25 | 49.59 | 1.56 | 1.51 | 1.49 | 1.56 | 1.50 | 1.54 | 30.99 | 23.42 | 47.66 | 35.05 | 26.25 | 49.59 | 1.60 | 1.10 | 1.29 | 1.66 | 1.09 | 1.31 | ZicoFullSTP | 11 |
| 12 | ZicoFullSTPFullData | 28.93 | 23.42 | 47.66 | 32.48 | 26.25 | 49.59 | 1.56 | 1.51 | 1.49 | 1.56 | 1.50 | 1.54 | 30.99 | 23.42 | 47.66 | 35.05 | 26.25 | 49.59 | 1.60 | 1.10 | 1.29 | 1.66 | 1.09 | 1.31 | ZicoFullSTPFullData | 12 |
| 13 | 3DShot1 | 28.72 | 22.80 | 18.68 | 32.71 | 24.30 | 23.19 | 1.51 | 1.30 | 1.61 | 0.93 | 28.72 | 22.80 | 18.68 | 32.71 | 24.30 | 23.19 | 1.05 | 0.88 | 1.15 | 0.47 | 3DShot1 | 13 | ||||
| 14 | RBO−Proteus | 28.51 | 22.73 | 47.29 | 32.24 | 25.70 | 49.07 | 1.46 | 1.28 | 1.45 | 1.50 | 1.34 | 1.48 | 28.72 | 24.73 | 48.03 | 32.48 | 27.96 | 49.86 | 1.05 | 1.57 | 1.33 | 1.10 | 1.64 | 1.35 | RBO−Proteus | 14 |
| 15 | Sasaki−Cetin−Sasai | 28.10 | 26.03 | 50.66 | 31.78 | 29.44 | 52.26 | 1.36 | 2.35 | 1.80 | 1.41 | 2.42 | 1.84 | 30.58 | 26.03 | 53.65 | 34.58 | 29.52 | 55.32 | 1.50 | 2.03 | 1.94 | 1.56 | 2.14 | 1.98 | Sasaki−Cetin−Sasai | 15 |
| 16 | Bates_BMM | 27.69 | 22.18 | 41.23 | 31.31 | 25.08 | 42.34 | 1.26 | 1.10 | 0.82 | 1.30 | 1.16 | 0.73 | 28.31 | 24.45 | 41.23 | 32.01 | 27.65 | 42.59 | 0.95 | 1.47 | 0.58 | 1.00 | 1.54 | 0.51 | Bates_BMM | 16 |
| 17 | MULTICOM | 27.48 | 21.97 | 41.51 | 31.07 | 24.84 | 42.77 | 1.21 | 1.04 | 0.85 | 1.25 | 1.09 | 0.78 | 31.20 | 24.38 | 46.72 | 35.05 | 27.34 | 48.46 | 1.66 | 1.44 | 1.18 | 1.66 | 1.44 | 1.18 | MULTICOM | 17 |
| 18 | POEMQA | 27.48 | 21.83 | 41.57 | 30.84 | 24.45 | 43.01 | 1.21 | 0.99 | 0.86 | 1.20 | 0.98 | 0.81 | 27.89 | 23.28 | 47.60 | 31.78 | 26.64 | 49.61 | 0.85 | 1.05 | 1.28 | 0.95 | 1.22 | 1.32 | POEMQA | 18 |
| 19 | FALCON_CONSENSUS | 27.07 | 22.38 | 42.97 | 28.74 | 23.44 | 44.50 | 1.11 | 1.17 | 1.00 | 0.75 | 0.68 | 0.97 | 29.96 | 24.73 | 46.74 | 33.41 | 27.96 | 48.40 | 1.35 | 1.57 | 1.19 | 1.30 | 1.64 | 1.18 | FALCON_CONSENSUS | 19 |
| 20 | HHpred2 | 26.86 | 22.86 | 29.39 | 30.14 | 25.62 | 33.17 | 1.06 | 1.32 | 1.05 | 1.32 | 26.86 | 22.86 | 29.39 | 30.14 | 25.62 | 33.17 | 0.60 | 0.90 | 0.59 | 0.89 | HHpred2 | 20 | ||||
| 21 | HHpred4 | 26.86 | 22.04 | 31.31 | 30.14 | 24.69 | 35.02 | 1.06 | 1.06 | 1.05 | 1.05 | 26.86 | 22.04 | 31.31 | 30.14 | 24.69 | 35.02 | 0.60 | 0.61 | 0.59 | 0.59 | HHpred4 | 21 | ||||
| 22 | HHpred5 | 26.86 | 22.04 | 31.31 | 30.14 | 24.69 | 35.02 | 1.06 | 1.06 | 1.05 | 1.05 | 26.86 | 22.04 | 31.31 | 30.14 | 24.69 | 35.02 | 0.60 | 0.61 | 0.59 | 0.59 | HHpred5 | 22 | ||||
| 23 | LEE | 26.65 | 21.07 | 43.78 | 29.91 | 23.60 | 45.27 | 1.01 | 0.74 | 1.09 | 1.00 | 0.73 | 1.06 | 29.96 | 22.38 | 45.65 | 33.88 | 25.31 | 47.31 | 1.35 | 0.73 | 1.07 | 1.41 | 0.79 | 1.05 | LEE | 23 |
| 24 | SHORTLE | 26.45 | 21.07 | 33.21 | 29.91 | 23.83 | 41.28 | 0.96 | 0.74 | 1.00 | 0.80 | 0.62 | 26.45 | 22.66 | 33.24 | 29.91 | 25.62 | 41.31 | 0.50 | 0.83 | 0.54 | 0.89 | 0.36 | SHORTLE | 24 | ||
| 25 | Handl−Lovell | 26.03 | 21.90 | 46.83 | 29.44 | 24.77 | 49.41 | 0.86 | 1.01 | 1.40 | 0.90 | 1.07 | 1.52 | 26.45 | 21.90 | 46.83 | 29.91 | 24.77 | 49.41 | 0.50 | 0.56 | 1.20 | 0.54 | 0.62 | 1.29 | Handl−Lovell | 25 |
| 26 | DBAKER | 26.03 | 19.56 | 42.72 | 29.44 | 22.12 | 44.86 | 0.86 | 0.25 | 0.98 | 0.90 | 0.30 | 1.01 | 26.45 | 20.80 | 42.72 | 29.44 | 22.12 | 44.86 | 0.50 | 0.17 | 0.74 | 0.43 | 0.77 | DBAKER | 26 | |
| 27 | POEM | 25.83 | 19.77 | 43.71 | 29.21 | 22.35 | 47.40 | 0.81 | 0.32 | 1.08 | 0.85 | 0.37 | 1.30 | 30.79 | 23.07 | 47.60 | 34.81 | 26.09 | 49.61 | 1.56 | 0.98 | 1.28 | 1.61 | 1.04 | 1.32 | POEM | 27 |
| 28 | IBT_LT | 25.62 | 23.55 | 25.30 | 28.97 | 26.64 | 33.07 | 0.76 | 1.55 | 0.80 | 1.61 | 25.62 | 23.55 | 25.30 | 28.97 | 26.64 | 33.07 | 0.29 | 1.15 | 0.33 | 1.22 | IBT_LT | 28 | ||||
| 29 | 3DShot2 | 25.62 | 22.25 | 32.06 | 29.67 | 25.86 | 33.58 | 0.76 | 1.13 | 0.95 | 1.39 | 25.62 | 22.25 | 32.06 | 29.67 | 25.86 | 33.58 | 0.29 | 0.69 | 0.48 | 0.97 | 3DShot2 | 29 | ||||
| 30 | FAMS−multi | 25.41 | 21.69 | 42.06 | 27.34 | 23.13 | 45.66 | 0.71 | 0.94 | 0.91 | 0.45 | 0.60 | 1.10 | 26.24 | 22.73 | 42.06 | 29.67 | 25.70 | 45.66 | 0.45 | 0.86 | 0.67 | 0.48 | 0.91 | 0.86 | FAMS−multi | 30 |
| 31 | COMA−M | 25.41 | 21.56 | 14.61 | 28.74 | 24.38 | 21.96 | 0.71 | 0.90 | 0.75 | 0.96 | 25.41 | 21.56 | 15.75 | 28.74 | 24.38 | 22.86 | 0.24 | 0.44 | 0.28 | 0.49 | COMA−M | 31 | ||||
| 32 | MUFOLD−MD | 25.41 | 20.73 | 42.18 | 28.50 | 23.21 | 43.52 | 0.71 | 0.63 | 0.92 | 0.70 | 0.62 | 0.87 | 25.41 | 20.73 | 42.18 | 28.50 | 23.21 | 43.52 | 0.24 | 0.14 | 0.69 | 0.23 | 0.12 | 0.61 | MUFOLD−MD | 32 |
| 33 | Elofsson | 25.00 | 22.52 | 34.23 | 28.27 | 25.31 | 35.87 | 0.61 | 1.21 | 0.10 | 0.65 | 1.23 | 0.02 | 25.21 | 23.28 | 35.56 | 28.27 | 25.31 | 36.40 | 0.19 | 1.05 | 0.17 | 0.79 | Elofsson | 33 | ||
| 34 | BAKER−ROBETTA | 24.79 | 17.91 | 44.13 | 28.04 | 20.25 | 46.30 | 0.56 | 1.12 | 0.60 | 1.17 | 31.20 | 23.35 | 44.13 | 35.05 | 26.64 | 46.30 | 1.66 | 1.08 | 0.90 | 1.66 | 1.22 | 0.93 | BAKER−ROBETTA | 34 | ||
| 35 | SAMUDRALA | 24.59 | 22.38 | 41.84 | 27.80 | 25.31 | 43.86 | 0.51 | 1.17 | 0.88 | 0.54 | 1.23 | 0.90 | 26.24 | 22.38 | 41.84 | 29.67 | 25.31 | 43.86 | 0.45 | 0.73 | 0.65 | 0.48 | 0.79 | 0.65 | SAMUDRALA | 35 |
| 36 | TJ_Jiang | 24.59 | 17.22 | 44.49 | 27.80 | 19.47 | 46.39 | 0.51 | 1.16 | 0.54 | 1.18 | 27.27 | 19.84 | 46.74 | 30.84 | 22.98 | 48.57 | 0.70 | 1.19 | 0.74 | 0.04 | 1.20 | TJ_Jiang | 36 | |||
| 37 | pro−sp3−TASSER | 24.38 | 20.80 | 35.97 | 27.34 | 23.29 | 39.04 | 0.46 | 0.66 | 0.28 | 0.45 | 0.64 | 0.37 | 24.38 | 20.80 | 42.02 | 27.34 | 23.29 | 44.00 | 0.17 | 0.67 | 0.14 | 0.67 | pro−sp3−TASSER | 37 | ||
| 38 | SAM−T08−server | 24.17 | 21.01 | 40.68 | 27.34 | 23.75 | 42.30 | 0.41 | 0.72 | 0.76 | 0.45 | 0.77 | 0.73 | 24.17 | 21.01 | 40.68 | 27.34 | 23.75 | 42.30 | 0.24 | 0.52 | 0.29 | 0.47 | SAM−T08−server | 38 | ||
| 39 | fais−server | 24.17 | 20.18 | 31.29 | 27.34 | 22.82 | 33.96 | 0.41 | 0.45 | 0.45 | 0.51 | 24.17 | 21.01 | 42.88 | 27.34 | 23.75 | 44.12 | 0.24 | 0.76 | 0.29 | 0.68 | fais−server | 39 | ||||
| 40 | fais@hgc | 24.17 | 20.18 | 31.29 | 27.34 | 22.82 | 33.96 | 0.41 | 0.45 | 0.45 | 0.51 | 24.17 | 20.18 | 43.66 | 27.34 | 22.82 | 45.09 | 0.85 | 0.79 | fais@hgc | 40 | ||||||
| 41 | McGuffin | 24.17 | 16.94 | 42.85 | 27.34 | 19.16 | 45.66 | 0.41 | 0.99 | 0.45 | 1.10 | 29.96 | 24.38 | 47.29 | 33.65 | 27.34 | 49.07 | 1.35 | 1.44 | 1.25 | 1.36 | 1.44 | 1.25 | McGuffin | 41 | ||
| 42 | MULTICOM−REFINE | 24.17 | 14.12 | 40.62 | 27.57 | 18.07 | 41.58 | 0.41 | 0.76 | 0.49 | 0.65 | 24.17 | 19.56 | 40.62 | 27.57 | 22.12 | 41.58 | 0.51 | 0.02 | 0.39 | MULTICOM−REFINE | 42 | |||||
| 43 | MUProt | 24.17 | 14.12 | 40.62 | 27.57 | 18.07 | 41.58 | 0.41 | 0.76 | 0.49 | 0.65 | 24.17 | 19.97 | 40.62 | 27.57 | 22.59 | 41.58 | 0.51 | 0.02 | 0.39 | MUProt | 43 | |||||
| 44 | PRI−Yang−KiharA | 23.97 | 21.35 | 43.52 | 27.10 | 24.14 | 45.23 | 0.36 | 0.83 | 1.06 | 0.39 | 0.89 | 1.06 | 23.97 | 21.35 | 43.52 | 27.10 | 24.14 | 45.23 | 0.37 | 0.83 | 0.41 | 0.81 | PRI−Yang−KiharA | 44 | ||
| 45 | pipe_int | 23.97 | 20.39 | 19.19 | 26.64 | 22.27 | 20.70 | 0.36 | 0.52 | 0.29 | 0.35 | 23.97 | 20.39 | 19.19 | 26.64 | 22.27 | 20.70 | 0.02 | pipe_int | 45 | |||||||
| 46 | JIVE08 | 23.97 | 17.08 | 43.47 | 27.10 | 19.32 | 45.63 | 0.36 | 1.05 | 0.39 | 1.10 | 23.97 | 17.29 | 43.47 | 27.10 | 19.55 | 45.63 | 0.83 | 0.86 | JIVE08 | 46 | ||||||
| 47 | GS−KudlatyPred | 23.76 | 20.39 | 41.13 | 26.64 | 22.82 | 42.68 | 0.31 | 0.52 | 0.81 | 0.29 | 0.51 | 0.77 | 28.93 | 23.83 | 41.13 | 32.71 | 26.95 | 42.97 | 1.10 | 1.25 | 0.57 | 1.15 | 1.32 | 0.55 | GS−KudlatyPred | 47 |
| 48 | Kolinski | 23.14 | 20.73 | 39.13 | 26.17 | 23.44 | 40.18 | 0.16 | 0.63 | 0.60 | 0.19 | 0.68 | 0.49 | 28.10 | 24.24 | 39.36 | 31.31 | 26.95 | 40.94 | 0.90 | 1.39 | 0.38 | 0.84 | 1.32 | 0.31 | Kolinski | 48 |
| 49 | Pcons_dot_net | 23.14 | 20.45 | 32.98 | 25.93 | 22.90 | 34.02 | 0.16 | 0.54 | 0.14 | 0.53 | 31.20 | 23.00 | 44.13 | 35.05 | 25.78 | 46.30 | 1.66 | 0.95 | 0.90 | 1.66 | 0.94 | 0.93 | Pcons_dot_net | 49 | ||
| 50 | Pcons_multi | 23.14 | 20.45 | 32.98 | 25.93 | 22.90 | 34.02 | 0.16 | 0.54 | 0.14 | 0.53 | 23.14 | 20.45 | 32.98 | 25.93 | 22.90 | 34.02 | 0.04 | 0.02 | Pcons_multi | 50 | ||||||
| 51 | SAINT1 | 22.93 | 19.08 | 43.75 | 24.77 | 20.33 | 45.26 | 0.11 | 0.10 | 1.08 | 1.06 | 22.93 | 19.08 | 43.75 | 24.77 | 20.33 | 45.26 | 0.86 | 0.81 | SAINT1 | 51 | ||||||
| 52 | FEIG | 22.73 | 19.77 | 31.24 | 25.70 | 22.35 | 34.76 | 0.06 | 0.32 | 0.09 | 0.37 | 26.86 | 23.97 | 39.59 | 29.21 | 25.93 | 41.62 | 0.60 | 1.30 | 0.40 | 0.38 | 0.99 | 0.39 | FEIG | 52 | ||
| 53 | 3D−JIGSAW_V3 | 22.73 | 18.87 | 17.86 | 25.70 | 21.34 | 24.32 | 0.06 | 0.03 | 0.09 | 0.08 | 23.97 | 21.07 | 22.14 | 27.10 | 23.83 | 28.78 | 0.27 | 0.31 | 3D−JIGSAW_V3 | 53 | ||||||
| 54 | GeneSilico | 22.52 | 20.87 | 26.77 | 25.70 | 23.83 | 30.72 | 0.01 | 0.68 | 0.09 | 0.80 | 25.83 | 20.87 | 42.38 | 29.21 | 23.83 | 44.43 | 0.35 | 0.19 | 0.71 | 0.38 | 0.31 | 0.72 | GeneSilico | 54 | ||
| 55 | LevittGroup | 22.52 | 19.56 | 42.35 | 25.93 | 22.59 | 43.07 | 0.01 | 0.25 | 0.94 | 0.14 | 0.44 | 0.82 | 30.58 | 24.11 | 47.77 | 34.58 | 27.26 | 48.64 | 1.50 | 1.35 | 1.30 | 1.56 | 1.41 | 1.20 | LevittGroup | 55 |
| 56 | PSI | 22.52 | 18.11 | 41.73 | 25.70 | 20.72 | 42.83 | 0.01 | 0.87 | 0.09 | 0.79 | 23.76 | 19.49 | 42.92 | 26.17 | 21.34 | 43.97 | 0.77 | 0.66 | PSI | 56 | ||||||
| 57 | CpHModels | 22.31 | 20.11 | 6.51 | 25.23 | 22.74 | 13.54 | 0.43 | 0.48 | 22.31 | 20.11 | 6.51 | 25.23 | 22.74 | 13.54 | CpHModels | 57 | ||||||||||
| 58 | MeilerLabRene | 22.31 | 19.42 | 32.13 | 25.23 | 21.96 | 33.29 | 0.21 | 0.26 | 22.31 | 19.42 | 33.11 | 25.23 | 21.96 | 35.19 | MeilerLabRene | 58 | ||||||||||
| 59 | MULTICOM−CLUSTER | 22.31 | 17.49 | 39.71 | 25.23 | 19.78 | 40.87 | 0.66 | 0.57 | 24.59 | 20.73 | 43.31 | 28.04 | 22.74 | 44.89 | 0.04 | 0.14 | 0.81 | 0.12 | 0.77 | MULTICOM−CLUSTER | 59 | |||||
| 60 | MUFOLD−Server | 22.11 | 17.70 | 33.17 | 25.47 | 20.48 | 35.20 | 0.04 | 22.11 | 17.70 | 33.91 | 25.47 | 20.48 | 36.00 | MUFOLD−Server | 60 | |||||||||||
| 61 | SMEG−CCP | 22.11 | 17.01 | 27.08 | 25.00 | 19.24 | 28.38 | 22.11 | 17.01 | 27.08 | 25.00 | 19.24 | 28.38 | SMEG−CCP | 61 | ||||||||||||
| 62 | METATASSER | 21.69 | 19.15 | 39.34 | 24.53 | 21.65 | 40.56 | 0.12 | 0.63 | 0.17 | 0.54 | 21.90 | 19.15 | 43.60 | 24.77 | 21.65 | 44.51 | 0.84 | 0.73 | METATASSER | 62 | ||||||
| 63 | POISE | 21.07 | 20.11 | 37.54 | 23.36 | 22.27 | 39.71 | 0.43 | 0.44 | 0.35 | 0.44 | 21.07 | 20.11 | 37.54 | 23.36 | 22.27 | 39.71 | 0.18 | 0.17 | POISE | 63 | ||||||
| 64 | TASSER | 21.07 | 17.22 | 33.27 | 23.83 | 19.78 | 35.85 | 0.01 | 24.59 | 18.94 | 38.26 | 27.80 | 21.73 | 40.54 | 0.04 | 0.26 | 0.07 | 0.27 | TASSER | 64 | |||||||
| 65 | Distill | 20.87 | 17.91 | 34.05 | 23.83 | 20.48 | 38.06 | 0.08 | 0.26 | 23.14 | 20.04 | 35.63 | 26.17 | 22.66 | 40.59 | 0.27 | Distill | 65 | |||||||||
| 66 | Pcons_local | 20.66 | 19.97 | 33.61 | 23.36 | 22.59 | 36.22 | 0.39 | 0.03 | 0.44 | 0.05 | 23.14 | 20.45 | 33.61 | 25.93 | 22.90 | 36.22 | 0.04 | 0.02 | Pcons_local | 66 | ||||||
| 67 | BioSerf | 20.66 | 18.18 | 38.62 | 23.36 | 20.56 | 40.32 | 0.55 | 0.51 | 27.89 | 21.69 | 41.57 | 31.54 | 24.53 | 42.29 | 0.85 | 0.49 | 0.62 | 0.89 | 0.54 | 0.47 | BioSerf | 67 | ||||
| 68 | FLOUDAS | 20.45 | 18.94 | 29.94 | 23.13 | 21.42 | 32.83 | 0.05 | 0.10 | 27.07 | 21.56 | 31.75 | 30.14 | 23.44 | 33.44 | 0.65 | 0.44 | 0.59 | 0.19 | FLOUDAS | 68 | ||||||
| 69 | RAPTOR | 20.45 | 17.29 | 35.13 | 23.60 | 20.02 | 36.50 | 0.19 | 0.09 | 22.73 | 20.52 | 43.47 | 25.93 | 22.98 | 45.30 | 0.07 | 0.83 | 0.04 | 0.82 | RAPTOR | 69 | ||||||
| 70 | 3DShotMQ | 20.45 | 16.12 | 28.14 | 23.13 | 18.22 | 29.36 | 20.45 | 16.12 | 28.14 | 23.13 | 18.22 | 29.36 | 3DShotMQ | 70 | ||||||||||||
| 71 | SAM−T06−server | 20.45 | 15.91 | 28.11 | 23.13 | 17.99 | 29.26 | 20.45 | 16.94 | 28.11 | 23.13 | 19.16 | 29.26 | SAM−T06−server | 71 | ||||||||||||
| 72 | Sternberg | 20.25 | 17.08 | 37.77 | 22.90 | 19.32 | 39.73 | 0.46 | 0.44 | 21.28 | 18.80 | 39.31 | 24.07 | 21.26 | 41.47 | 0.37 | 0.38 | Sternberg | 72 | ||||||||
| 73 | Hao_Kihara | 20.04 | 17.77 | 28.97 | 22.66 | 20.25 | 30.57 | 20.04 | 17.77 | 28.97 | 22.66 | 20.25 | 30.57 | Hao_Kihara | 73 | ||||||||||||
| 74 | FAMSD | 20.04 | 16.60 | 31.52 | 22.66 | 18.77 | 33.32 | 21.07 | 18.32 | 31.52 | 24.07 | 20.17 | 33.32 | FAMSD | 74 | ||||||||||||
| 75 | Bilab−UT | 19.84 | 18.59 | 38.84 | 22.43 | 21.03 | 40.24 | 0.57 | 0.50 | 26.86 | 21.07 | 43.50 | 30.37 | 23.83 | 44.59 | 0.60 | 0.27 | 0.83 | 0.64 | 0.31 | 0.74 | Bilab−UT | 75 | ||||
| 76 | Wolynes | 19.84 | 17.49 | 30.24 | 22.43 | 19.47 | 31.79 | 19.84 | 17.49 | 34.28 | 22.43 | 19.47 | 35.91 | Wolynes | 76 | ||||||||||||
| 77 | MULTICOM−RANK | 19.63 | 16.18 | 27.29 | 21.73 | 17.84 | 29.86 | 22.52 | 19.90 | 32.61 | 25.47 | 22.51 | 33.34 | MULTICOM−RANK | 77 | ||||||||||||
| 78 | Frankenstein | 19.42 | 17.77 | 24.68 | 21.96 | 20.09 | 26.23 | 23.35 | 20.52 | 34.19 | 26.64 | 22.51 | 34.99 | 0.07 | Frankenstein | 78 | |||||||||||
| 79 | MidwayFolding | 19.21 | 16.74 | 32.93 | 21.96 | 19.16 | 35.91 | 0.02 | 19.42 | 16.94 | 33.88 | 21.96 | 19.16 | 35.91 | MidwayFolding | 79 | |||||||||||
| 80 | EB_AMU_Physics | 18.80 | 15.22 | 31.70 | 20.56 | 17.06 | 32.61 | 18.80 | 15.22 | 31.70 | 20.56 | 17.06 | 32.61 | EB_AMU_Physics | 80 | ||||||||||||
| 81 | Chicken_George | 18.80 | 15.08 | 41.82 | 21.26 | 17.06 | 45.08 | 0.88 | 1.04 | 24.79 | 20.39 | 42.09 | 26.87 | 23.05 | 45.36 | 0.09 | 0.02 | 0.68 | 0.06 | 0.83 | Chicken_George | 81 | |||||
| 82 | 3D−JIGSAW_AEP | 18.39 | 17.84 | 24.40 | 20.79 | 20.17 | 29.21 | 25.41 | 20.52 | 30.31 | 28.50 | 22.90 | 37.36 | 0.24 | 0.07 | 0.23 | 0.02 | 3D−JIGSAW_AEP | 82 | ||||||||
| 83 | DistillSN | 18.39 | 16.87 | 19.07 | 20.79 | 19.08 | 22.58 | 19.63 | 16.87 | 24.55 | 22.20 | 19.24 | 28.20 | DistillSN | 83 | ||||||||||||
| 84 | LEE−SERVER | 18.39 | 16.74 | 31.88 | 20.79 | 18.93 | 32.73 | 19.01 | 16.74 | 31.88 | 21.73 | 18.93 | 32.73 | LEE−SERVER | 84 | ||||||||||||
| 85 | mariner1 | 18.39 | 15.50 | 9.74 | 20.79 | 17.52 | 16.90 | 20.87 | 17.49 | 21.33 | 23.60 | 19.78 | 23.72 | mariner1 | 85 | ||||||||||||
| 86 | Jiang_Zhu | 18.18 | 15.70 | 26.72 | 20.79 | 17.37 | 29.26 | 18.18 | 15.70 | 26.72 | 20.79 | 17.37 | 29.26 | Jiang_Zhu | 86 | ||||||||||||
| 87 | Phyre_de_novo | 18.18 | 15.56 | 25.39 | 20.56 | 17.60 | 27.00 | 19.21 | 16.32 | 31.74 | 21.96 | 19.00 | 33.25 | Phyre_de_novo | 87 | ||||||||||||
| 88 | mGenTHREADER | 17.98 | 16.67 | 30.44 | 20.33 | 18.85 | 37.34 | 0.18 | 17.98 | 16.67 | 30.44 | 20.33 | 18.85 | 37.34 | mGenTHREADER | 88 | |||||||||||
| 89 | 3Dpro | 17.98 | 15.29 | 28.71 | 20.09 | 17.13 | 31.46 | 17.98 | 16.12 | 28.71 | 20.09 | 17.45 | 31.46 | 3Dpro | 89 | ||||||||||||
| 90 | FOLDpro | 17.98 | 15.29 | 27.34 | 20.09 | 16.51 | 30.00 | 19.21 | 15.29 | 27.34 | 21.96 | 16.98 | 30.00 | FOLDpro | 90 | ||||||||||||
| 91 | Scheraga | 17.77 | 15.77 | 31.51 | 20.09 | 17.84 | 32.50 | 25.41 | 19.77 | 39.60 | 28.74 | 22.35 | 40.26 | 0.24 | 0.40 | 0.28 | 0.24 | Scheraga | 91 | ||||||||
| 92 | nFOLD3 | 17.77 | 14.67 | 32.38 | 20.09 | 16.59 | 33.42 | 23.76 | 20.32 | 32.38 | 24.53 | 22.35 | 33.42 | nFOLD3 | 92 | ||||||||||||
| 93 | GS−MetaServer2 | 17.56 | 17.29 | 31.38 | 19.86 | 19.55 | 33.13 | 22.31 | 18.59 | 31.38 | 25.23 | 21.03 | 33.13 | GS−MetaServer2 | 93 | ||||||||||||
| 94 | GeneSilicoMetaServer | 17.56 | 17.29 | 31.38 | 19.86 | 19.55 | 33.13 | 22.31 | 18.59 | 31.38 | 25.23 | 21.03 | 33.13 | GeneSilicoMetaServer | 94 | ||||||||||||
| 95 | Phragment | 17.56 | 15.63 | 26.08 | 19.86 | 17.68 | 27.96 | 19.63 | 17.42 | 28.02 | 22.20 | 19.70 | 30.23 | Phragment | 95 | ||||||||||||
| 96 | Poing | 17.56 | 14.39 | 20.70 | 19.86 | 16.28 | 24.03 | 17.56 | 15.98 | 24.13 | 19.86 | 18.07 | 26.69 | Poing | 96 | ||||||||||||
| 97 | Phyre2 | 17.36 | 16.39 | 19.88 | 18.93 | 15.65 | 23.59 | 20.25 | 19.28 | 29.12 | 22.90 | 21.81 | 33.30 | Phyre2 | 97 | ||||||||||||
| 98 | SAM−T02−server | 17.36 | 15.56 | 7.53 | 19.63 | 17.60 | 14.61 | 17.36 | 15.56 | 7.66 | 19.63 | 17.60 | 14.74 | SAM−T02−server | 98 | ||||||||||||
| 99 | ACOMPMOD | 17.36 | 15.01 | 16.73 | 19.63 | 16.98 | 17.40 | 18.59 | 15.70 | 21.48 | 21.03 | 17.52 | 22.76 | ACOMPMOD | 99 | ||||||||||||
| 100 | PS2−server | 17.36 | 14.12 | 26.86 | 18.69 | 14.49 | 29.74 | 22.52 | 18.80 | 35.80 | 25.47 | 21.26 | 38.82 | 0.07 | PS2−server | 100 | |||||||||||
| 101 | PS2−manual | 17.36 | 14.12 | 26.86 | 18.69 | 14.49 | 29.74 | 22.52 | 18.80 | 35.80 | 25.47 | 21.26 | 38.82 | 0.07 | PS2−manual | 101 | |||||||||||
| 102 | FFASstandard | 17.36 | 11.71 | 19.63 | 13.24 | 5.32 | 17.36 | 14.46 | 19.63 | 16.36 | 6.14 | FFASstandard | 102 | ||||||||||||||
| 103 | FFASflextemplate | 17.36 | 11.71 | 19.63 | 13.24 | 5.32 | 17.36 | 14.46 | 19.63 | 16.36 | 6.14 | FFASflextemplate | 103 | ||||||||||||||
| 104 | RANDOM | 17.30 | 14.39 | 17.30 | 20.29 | 16.74 | 20.29 | 17.30 | 14.39 | 17.30 | 20.29 | 16.74 | 20.29 | RANDOM | 104 | ||||||||||||
| 105 | circle | 17.15 | 16.05 | 28.19 | 19.39 | 18.15 | 32.01 | 19.84 | 16.80 | 34.49 | 22.66 | 20.72 | 36.35 | circle | 105 | ||||||||||||
| 106 | MULTICOM−CMFR | 17.15 | 15.29 | 22.78 | 19.39 | 17.29 | 24.56 | 22.73 | 15.98 | 31.93 | 25.70 | 18.07 | 34.08 | MULTICOM−CMFR | 106 | ||||||||||||
| 107 | A−TASSER | 16.94 | 14.81 | 37.09 | 18.93 | 16.51 | 39.70 | 0.39 | 0.44 | 20.87 | 18.87 | 37.09 | 23.60 | 21.34 | 39.70 | 0.13 | 0.17 | A−TASSER | 107 | ||||||||
| 108 | keasar−server | 16.74 | 15.56 | 17.21 | 18.93 | 17.60 | 19.19 | 16.74 | 15.56 | 17.21 | 18.93 | 17.60 | 19.19 | keasar−server | 108 | ||||||||||||
| 109 | MUSTER | 16.74 | 14.26 | 20.75 | 18.69 | 14.02 | 23.43 | 21.28 | 18.18 | 35.05 | 24.30 | 20.95 | 36.33 | MUSTER | 109 | ||||||||||||
| 110 | forecast | 16.53 | 15.56 | 20.70 | 18.69 | 17.45 | 21.52 | 18.39 | 17.01 | 20.70 | 20.33 | 19.08 | 21.52 | forecast | 110 | ||||||||||||
| 111 | Softberry | 16.53 | 14.88 | 22.44 | 18.69 | 16.82 | 23.38 | 17.98 | 17.15 | 31.58 | 20.33 | 19.39 | 33.14 | Softberry | 111 | ||||||||||||
| 112 | FFASsuboptimal | 16.32 | 13.29 | 18.46 | 15.03 | 2.78 | 17.77 | 17.29 | 20.09 | 19.55 | 6.20 | FFASsuboptimal | 112 | ||||||||||||||
| 113 | schenk−torda−server | 16.12 | 13.77 | 21.03 | 18.22 | 15.58 | 23.48 | 16.12 | 14.95 | 21.03 | 18.22 | 16.90 | 23.48 | schenk−torda−server | 113 | ||||||||||||
| 114 | COMA | 16.12 | 12.81 | 11.93 | 18.22 | 14.49 | 19.18 | 25.41 | 21.56 | 15.75 | 28.74 | 24.38 | 22.86 | 0.24 | 0.44 | 0.28 | 0.49 | COMA | 114 | ||||||||
| 115 | StruPPi | 15.29 | 14.19 | 15.40 | 17.52 | 15.34 | 18.61 | 15.29 | 14.19 | 15.40 | 17.52 | 15.34 | 18.61 | StruPPi | 115 | ||||||||||||
| 116 | mahmood−torda−server | 15.08 | 11.91 | 22.81 | 17.06 | 13.47 | 24.74 | 16.12 | 14.74 | 22.81 | 18.22 | 16.51 | 24.74 | mahmood−torda−server | 116 | ||||||||||||
| 117 | TWPPLAB | 13.64 | 11.71 | 5.24 | 15.42 | 13.24 | 6.15 | 13.64 | 11.71 | 5.24 | 15.42 | 13.24 | 6.15 | TWPPLAB | 117 | ||||||||||||
| 118 | fleil | 12.40 | 11.02 | 8.93 | 14.02 | 12.46 | 9.80 | 14.46 | 13.91 | 9.33 | 16.36 | 15.73 | 10.14 | fleil | 118 | ||||||||||||
| 119 | OLGAFS | 12.40 | 10.81 | 14.02 | 12.23 | 14.46 | 13.91 | 16.36 | 15.73 | OLGAFS | 119 | ||||||||||||||||
| 120 | rehtnap | 9.09 | 8.13 | 10.28 | 9.19 | 9.50 | 9.23 | 10.75 | 10.44 | rehtnap | 120 | ||||||||||||||||
| 121 | huber−torda−server | 0.00 | 0.00 | 0.00 | 0.00 | 17.56 | 13.43 | 12.43 | 19.86 | 15.19 | 15.14 | huber−torda−server | 121 | ||||||||||||||
| 122 | Pushchino | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | Pushchino | 122 | ||||||||||||||||
| 123 | FUGUE_KM | 0.00 | 0.00 | 0.00 | 0.00 | 15.29 | 13.57 | 5.73 | 17.29 | 15.34 | 8.21 | FUGUE_KM | 123 | ||||||||||||||
| 124 | panther_server | 0.00 | 0.00 | 0.00 | 0.00 | 12.81 | 11.43 | 13.55 | 10.75 | panther_server | 124 | ||||||||||||||||
| 125 | AMU−Biology | AMU−Biology | 125 | ||||||||||||||||||||||||
| 126 | Abagyan | Abagyan | 126 | ||||||||||||||||||||||||
| 127 | BHAGEERATH | BHAGEERATH | 127 | ||||||||||||||||||||||||
| 128 | CBSU | CBSU | 128 | ||||||||||||||||||||||||
| 129 | DelCLab | DelCLab | 129 | ||||||||||||||||||||||||
| 130 | FEIG_REFINE | FEIG_REFINE | 130 | ||||||||||||||||||||||||
| 131 | Fiser−M4T | Fiser−M4T | 131 | ||||||||||||||||||||||||
| 132 | FrankensteinLong | FrankensteinLong | 132 | ||||||||||||||||||||||||
| 133 | HCA | HCA | 133 | ||||||||||||||||||||||||
| 134 | KudlatyPredHuman | KudlatyPredHuman | 134 | ||||||||||||||||||||||||
| 135 | LOOPP_Server | LOOPP_Server | 135 | ||||||||||||||||||||||||
| 136 | Linnolt−UH−CMB | Linnolt−UH−CMB | 136 | ||||||||||||||||||||||||
| 137 | NIM2 | NIM2 | 137 | ||||||||||||||||||||||||
| 138 | Nano_team | Nano_team | 138 | ||||||||||||||||||||||||
| 139 | NirBenTal | NirBenTal | 139 | ||||||||||||||||||||||||
| 140 | Ozkan−Shell | Ozkan−Shell | 140 | ||||||||||||||||||||||||
| 141 | PHAISTOS | PHAISTOS | 141 | ||||||||||||||||||||||||
| 142 | PZ−UAM | PZ−UAM | 142 | ||||||||||||||||||||||||
| 143 | ProtAnG | ProtAnG | 143 | ||||||||||||||||||||||||
| 144 | ProteinShop | ProteinShop | 144 | ||||||||||||||||||||||||
| 145 | RPFM | RPFM | 145 | ||||||||||||||||||||||||
| 146 | ShakAbInitio | ShakAbInitio | 146 | ||||||||||||||||||||||||
| 147 | TsaiLab | TsaiLab | 147 | ||||||||||||||||||||||||
| 148 | UCDavisGenome | UCDavisGenome | 148 | ||||||||||||||||||||||||
| 149 | Wolfson−FOBIA | Wolfson−FOBIA | 149 | ||||||||||||||||||||||||
| 150 | YASARA | YASARA | 150 | ||||||||||||||||||||||||
| 151 | YASARARefine | YASARARefine | 151 | ||||||||||||||||||||||||
| 152 | Zhou−SPARKS | Zhou−SPARKS | 152 | ||||||||||||||||||||||||
| 153 | dill_ucsf | dill_ucsf | 153 | ||||||||||||||||||||||||
| 154 | dill_ucsf_extended | dill_ucsf_extended | 154 | ||||||||||||||||||||||||
| 155 | igor | igor | 155 | ||||||||||||||||||||||||
| 156 | jacobson | jacobson | 156 | ||||||||||||||||||||||||
| 157 | mti | mti | 157 | ||||||||||||||||||||||||
| 158 | mumssp | mumssp | 158 | ||||||||||||||||||||||||
| 159 | psiphifoldings | psiphifoldings | 159 | ||||||||||||||||||||||||
| 160 | ricardo | ricardo | 160 | ||||||||||||||||||||||||
| 161 | rivilo | rivilo | 161 | ||||||||||||||||||||||||
| 162 | sessions | sessions | 162 | ||||||||||||||||||||||||
| 163 | taylor | taylor | 163 | ||||||||||||||||||||||||
| 164 | test_http_server_01 | test_http_server_01 | 164 | ||||||||||||||||||||||||
| 165 | tripos_08 | tripos_08 | 165 | ||||||||||||||||||||||||
| 166 | xianmingpan | xianmingpan | 166 | ||||||||||||||||||||||||