GO annotation search and functional categorization
Among 46147 Citrus sinensis proteins, 32986 are assigned at least on GO term. Predicted GO terms with confidence scores greater than 10 points, 8 points and 6 points are considered to be 'very confident', 'confident' and 'probable', respectively. We further mapped the predicted GO terms to Plant GO slim, which presents a general categorization of GO content and is useful for the broad protein function classification.
The functional categorization listing is shown below. Clicking on the number of GO annotation count navigates to the page that shows the list of predicted GO terms associated with corresponding GO slim term
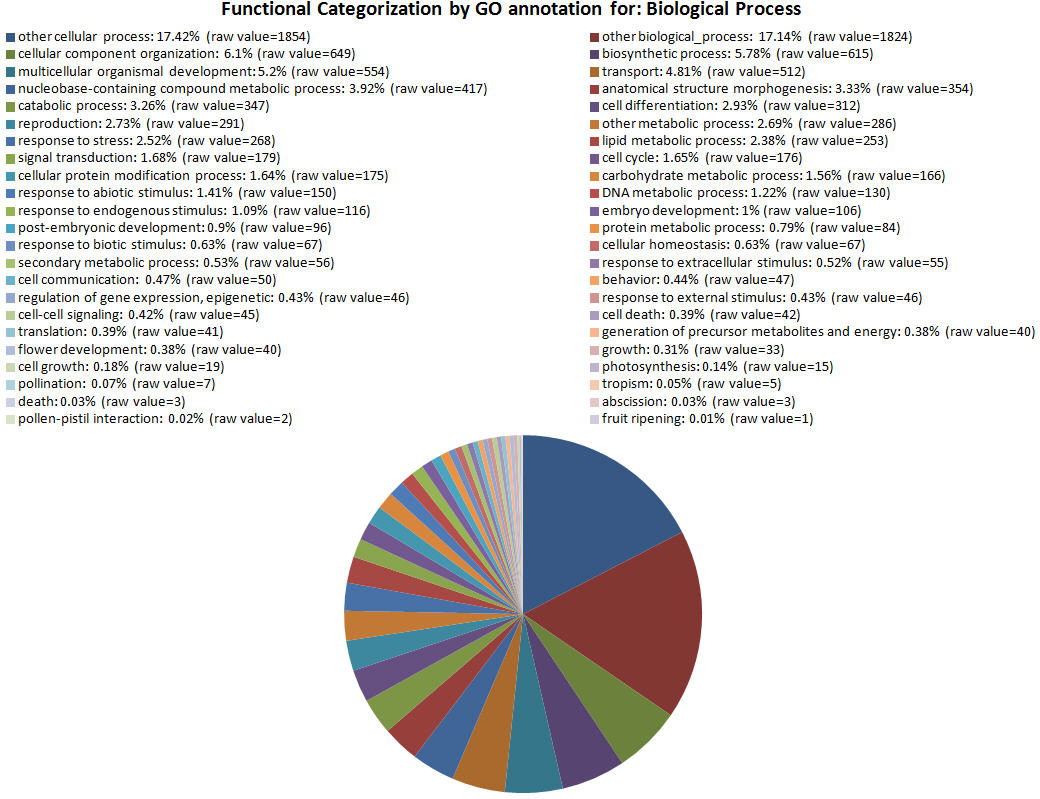
| GOID Link to GO |
Functional Category | GO Annotation Count to see GO terms |
|---|---|---|
| GO:0009987 | other cellular process | 1854 |
| GO:0008150 | other biological_process | 1824 |
| GO:0016043 | cellular component organization | 649 |
| GO:0009058 | biosynthetic process | 615 |
| GO:0007275 | multicellular organismal development | 554 |
| GO:0006810 | transport | 512 |
| GO:0006139 | nucleobase-containing compound metabolic process | 417 |
| GO:0009653 | anatomical structure morphogenesis | 354 |
| GO:0009056 | catabolic process | 347 |
| GO:0030154 | cell differentiation | 312 |
| GO:0000003 | reproduction | 291 |
| GO:0008152 | other metabolic process | 286 |
| GO:0006950 | response to stress | 268 |
| GO:0006629 | lipid metabolic process | 253 |
| GO:0007165 | signal transduction | 179 |
| GO:0007049 | cell cycle | 176 |
| GO:0006464 | cellular protein modification process | 175 |
| GO:0005975 | carbohydrate metabolic process | 166 |
| GO:0009628 | response to abiotic stimulus | 150 |
| GO:0006259 | DNA metabolic process | 130 |
| GO:0009719 | response to endogenous stimulus | 116 |
| GO:0009790 | embryo development | 106 |
| GO:0009791 | post-embryonic development | 96 |
| GO:0019538 | protein metabolic process | 84 |
| GO:0009607 | response to biotic stimulus | 67 |
| GO:0019725 | cellular homeostasis | 67 |
| GO:0019748 | secondary metabolic process | 56 |
| GO:0009991 | response to extracellular stimulus | 55 |
| GO:0007154 | cell communication | 50 |
| GO:0007610 | behavior | 47 |
| GO:0040029 | regulation of gene expression, epigenetic | 46 |
| GO:0009605 | response to external stimulus | 46 |
| GO:0007267 | cell-cell signaling | 45 |
| GO:0008219 | cell death | 42 |
| GO:0006412 | translation | 41 |
| GO:0006091 | generation of precursor metabolites and energy | 40 |
| GO:0009908 | flower development | 40 |
| GO:0040007 | growth | 33 |
| GO:0016049 | cell growth | 19 |
| GO:0015979 | photosynthesis | 15 |
| GO:0009856 | pollination | 7 |
| GO:0009606 | tropism | 5 |
| GO:0016265 | death | 3 |
| GO:0009838 | abscission | 3 |
| GO:0009875 | pollen-pistil interaction | 2 |
| GO:0009835 | fruit ripening | 1 |
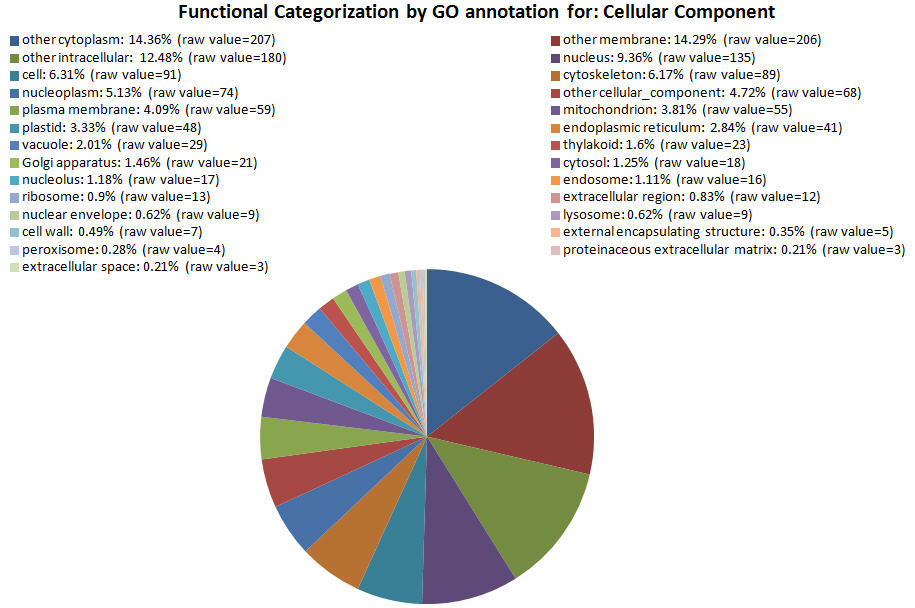
| GOID Link to GO |
Functional Category | GO Annotation Count to see GO terms |
|---|---|---|
| GO:0005737 | other cytoplasm | 207 |
| GO:0016020 | other membrane | 206 |
| GO:0005622 | other intracellular | 180 |
| GO:0005634 | nucleus | 135 |
| GO:0005623 | cell | 91 |
| GO:0005856 | cytoskeleton | 89 |
| GO:0005654 | nucleoplasm | 74 |
| GO:0005575 | other cellular_component | 68 |
| GO:0005886 | plasma membrane | 59 |
| GO:0005739 | mitochondrion | 55 |
| GO:0009536 | plastid | 48 |
| GO:0005783 | endoplasmic reticulum | 41 |
| GO:0005773 | vacuole | 29 |
| GO:0009579 | thylakoid | 23 |
| GO:0005794 | Golgi apparatus | 21 |
| GO:0005829 | cytosol | 18 |
| GO:0005730 | nucleolus | 17 |
| GO:0005768 | endosome | 16 |
| GO:0005840 | ribosome | 13 |
| GO:0005576 | extracellular region | 12 |
| GO:0005635 | nuclear envelope | 9 |
| GO:0005764 | lysosome | 9 |
| GO:0005618 | cell wall | 7 |
| GO:0030312 | external encapsulating structure | 5 |
| GO:0005777 | peroxisome | 4 |
| GO:0005578 | proteinaceous extracellular matrix | 3 |
| GO:0005615 | extracellular space | 3 |
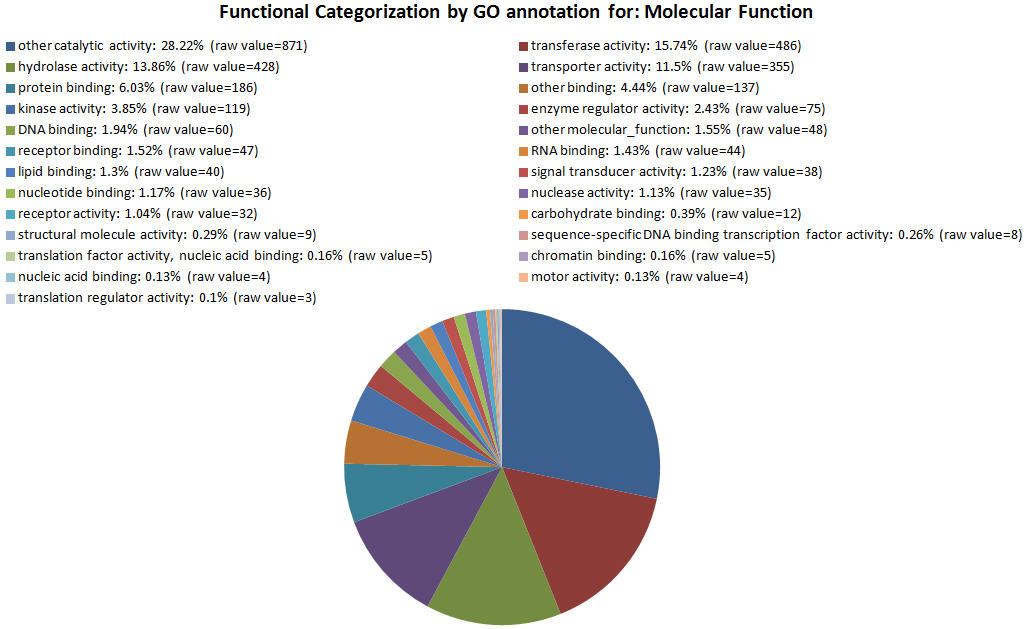
| GOID Link to GO |
Functional Category | GO Annotation Count to see GO terms |
|---|---|---|
| GO:0003824 | other catalytic activity | 871 |
| GO:0016740 | transferase activity | 486 |
| GO:0016787 | hydrolase activity | 428 |
| GO:0005215 | transporter activity | 355 |
| GO:0005515 | protein binding | 186 |
| GO:0005488 | other binding | 137 |
| GO:0016301 | kinase activity | 119 |
| GO:0030234 | enzyme regulator activity | 75 |
| GO:0003677 | DNA binding | 60 |
| GO:0003674 | other molecular_function | 48 |
| GO:0005102 | receptor binding | 47 |
| GO:0003723 | RNA binding | 44 |
| GO:0008289 | lipid binding | 40 |
| GO:0004871 | signal transducer activity | 38 |
| GO:0000166 | nucleotide binding | 36 |
| GO:0004518 | nuclease activity | 35 |
| GO:0004872 | receptor activity | 32 |
| GO:0030246 | carbohydrate binding | 12 |
| GO:0005198 | structural molecule activity | 9 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 8 |
| GO:0008135 | translation factor activity, nucleic acid binding | 5 |
| GO:0003682 | chromatin binding | 5 |
| GO:0003676 | nucleic acid binding | 4 |
| GO:0003774 | motor activity | 4 |
| GO:0045182 | translation regulator activity | 3 |