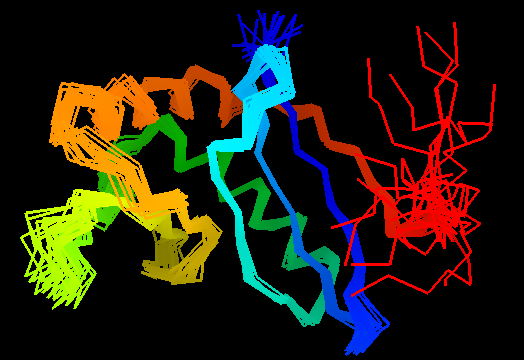
T0472
UPF0339 protein SO3888 from Shewanella oneidensis
Target sequence:
>T0472 SoR190, Shewanella oneidensis MR-1, 110 residues
MSGWYELSKSSNDQFKFVLKAGNGEVILTSELYTGKSGAMNGIESVQTNSPIEARYAKEVAKNDKPYFNLKAANHQIIGTSQMYSSTAARDNGIKSVMENGKTTTIKDLT
Structure:
Method: NMR
Determined by: NESG
PDB ID: 2k49
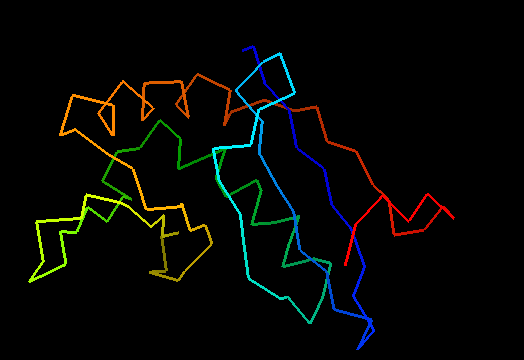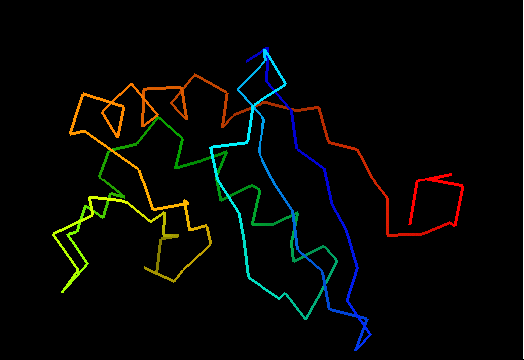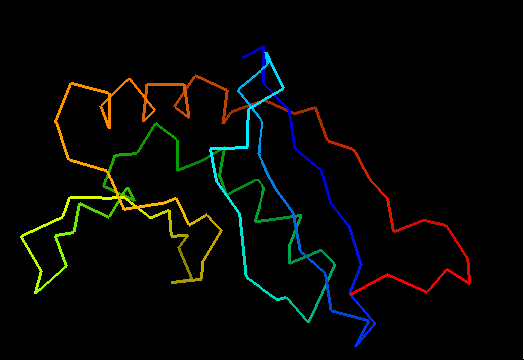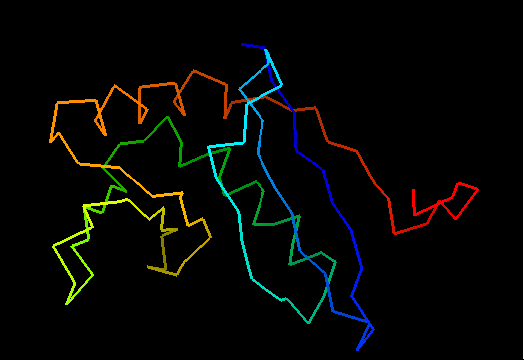
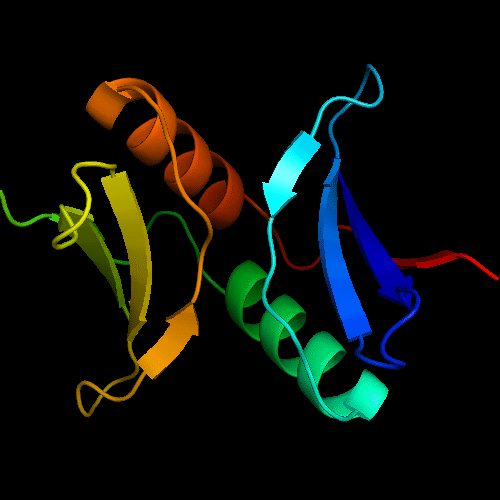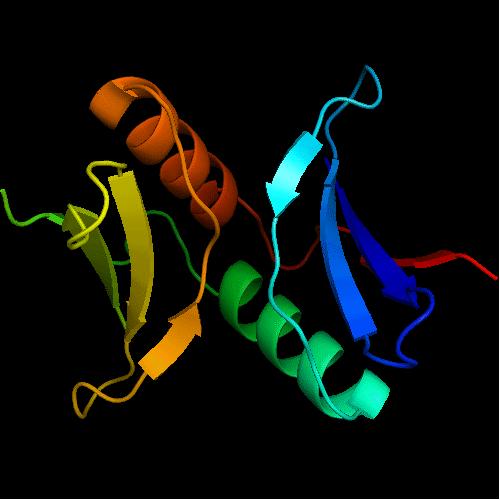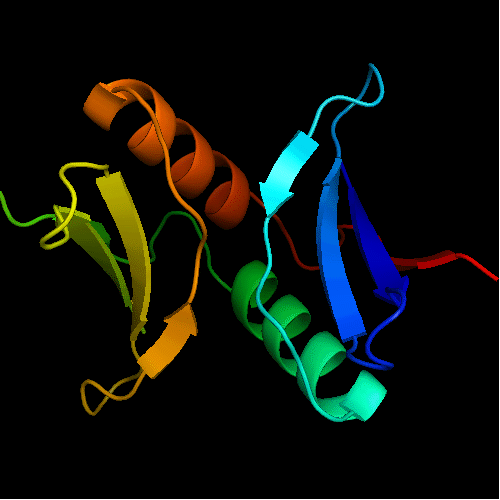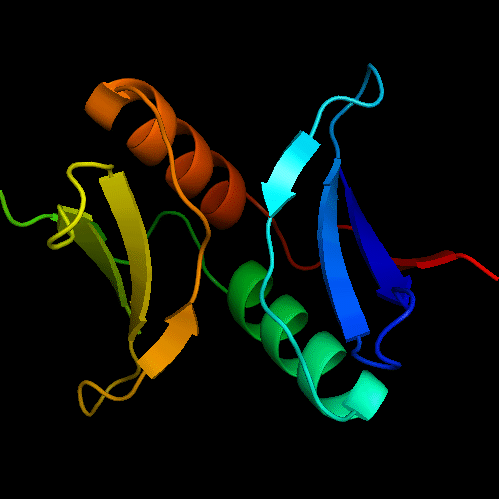
Domains: PyMOL of domains
Duplication: two domains. Domain 1: 1-50 and 102-110. Domain 2: 51-101. For this target, evaluation of the whole 2-domain structure is particularly interesting, as the main challenge was in relative positioning of the domains. Poorly structured N- and C-termini. Structurally variable regions were removed from NMR models for evaluation of predictions.
Domains of T0472: 2k49 model 1 residues 2-110
N- and C-domains are colored blue and red
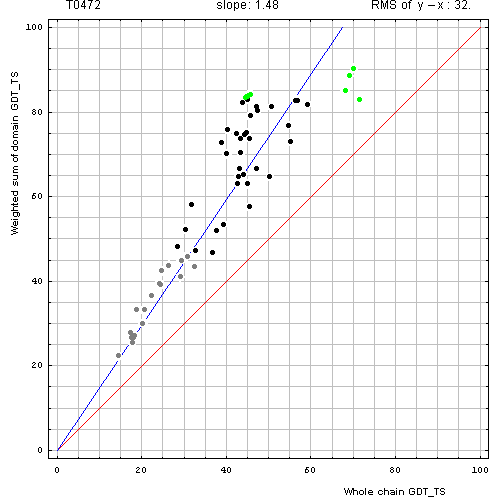
Correlation between weighted by the number of residues sum of GDT-TS scores for domain-based evlatuation (y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
To compute the weighted sum, GDT-TS for each domain was multiplied by the domain length, and this sum was divided by the sum of domain lengths. Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the weighted sum of domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
Tandem duplication of a β3α unit similar to DNA-binding domain, or dsRNA-binding domain (dsRBD) without the first α-helix. Domain-swapped with an additional C-terminal β-strand. This β-strand completes the fold in a manner similar to that in the shorter variants of RNAse H-like fold, e.g. Ada DNA-repair domain.
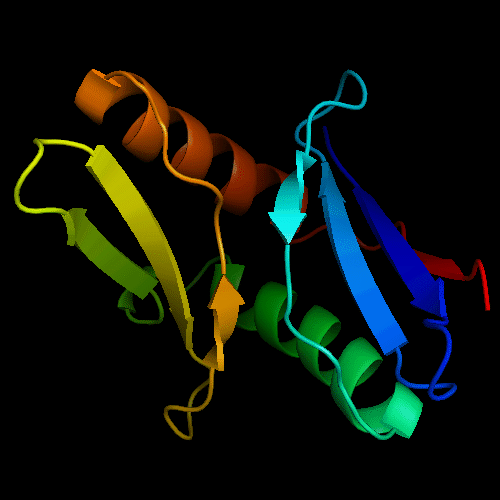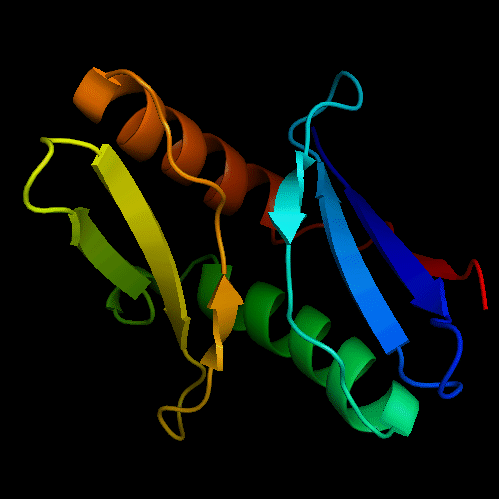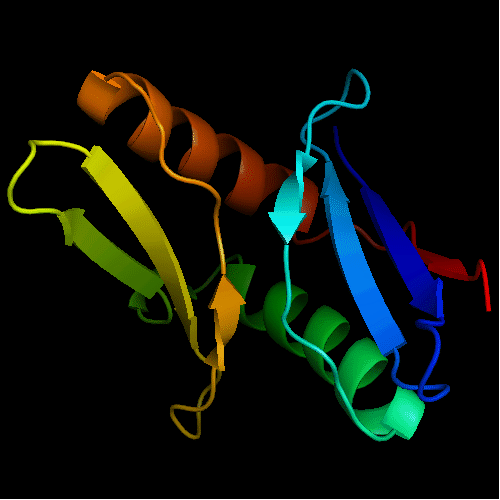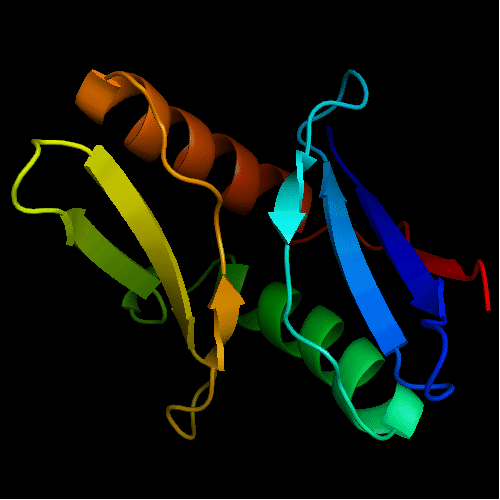
Cartoon diagram of T0472: 2k49 model 1 residues 2-110
CASP category:
Full chain: Comparative modeling: hard. Closest templates detectable by PSI-BLAST on the second iteration, but relative positioning of the duplicates is a problem.
For individual domains: Comparative modeling: easy.
Closest templates:
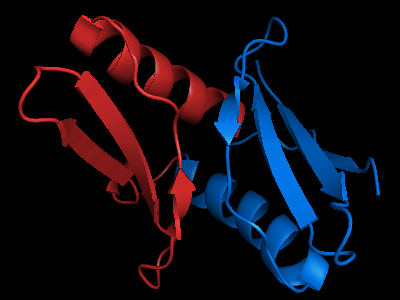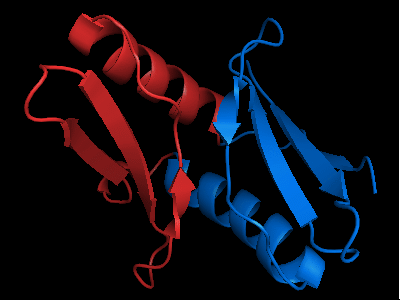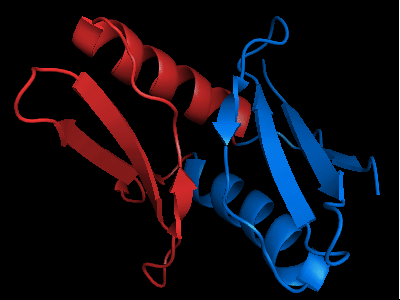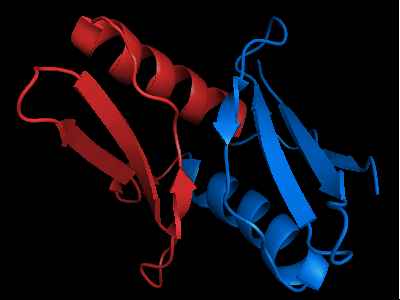
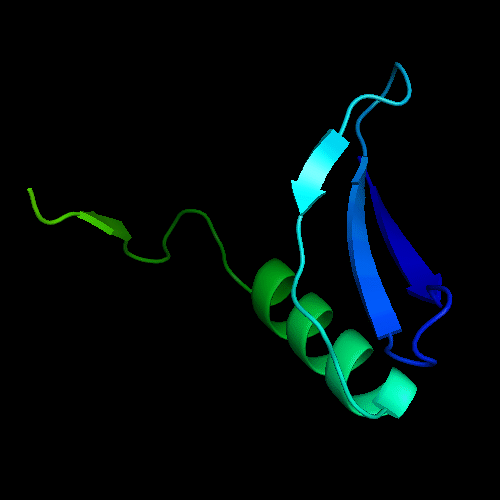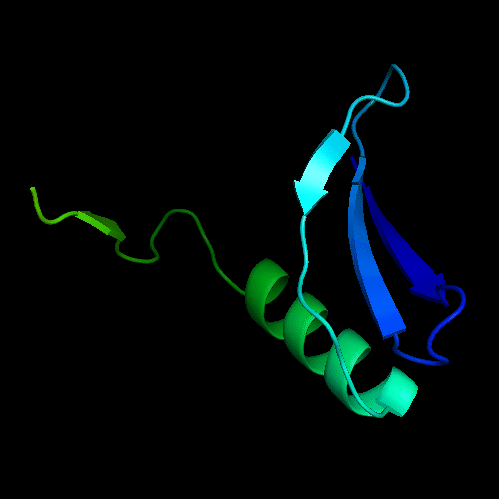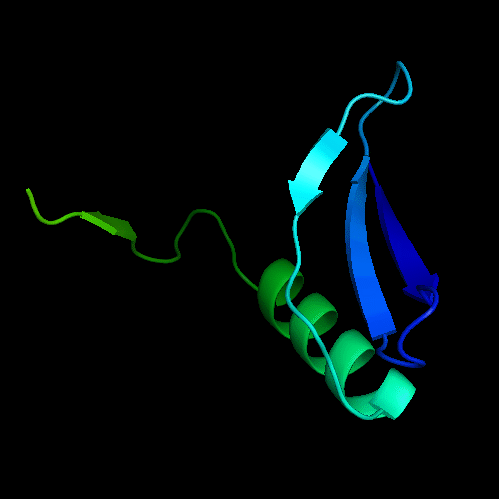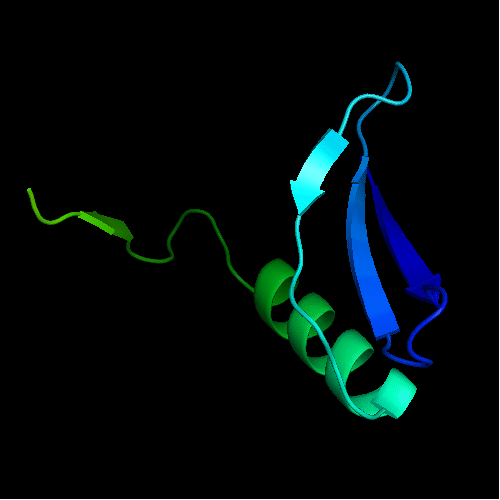
The only available template is NMB1088 protein from Neisseria meningitidis: 3bid, which is a domain-swapped dimer. The whole dimer should be used for the tandemly duplicated T0472 sequence. Interestingly, to close the sequence between the duplicates, one β-strand (last strand in chain A, green, after the α-helix) needs to be deleted from the dimeric template composed of chains A and B.
Cartoon diagram of the dimeric template: 3bid chains A (blue-green) and B (lime-red)
Chain A of the template 3bid
Duplicates of T0473 are aligned to 3bid. Magenta pairs form ionic interactions (guess which one with which):
T0472_1 MSGWYELSKSSNDQFKFVLKAGNGEVILTSELYTGKSGAMNGIESVQTNSPIEA------
T0472_2 ---RYAKEVAKNDKPYFNLKAANHQIIGTSQMYSSTAARDNGIKSVMENGKTTTIKDLT-
3bid_A --MYFEIYKDAKGEYRWRLKAANHEIIAQGEGYTSKQNCQHAVDLLKSTTAATPVKEVLE
Target sequence - PDB file inconsistencies:
NMR models contain C-terminal His-tag residues 111-LEHHHHHH-118 not present in the target sequence. Majority of this region was removed from NMR models due to disorder anyway.
T0472 2k49.pdb T0472.pdb PyMOL PyMOL of domains
T0472 1 MSGWYELSKSSNDQFKFVLKAGNGEVILTSELYTGKSGAMNGIESVQTNSPIEARYAKEVAKNDKPYFNLKAANHQIIGTSQMYSSTAARDNGIKSVMENGKTTTIKDLT-------- 110 *|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~ 2k49A 1 MSGWYELSKSSNDQFKFVLKAGNGEVILTSELYTGKSGAMNGIESVQTNSPIEARYAKEVAKNDKPYFNLKAANHQIIGTSQMYSSTAARDNGIKSVMENGKTTTIKDLTLEHHHHHH 118
Residue change log: remove 111-LEHHHHHH-118 as it is not present in target sequence; remove 1 M by 3.5Å SD cutoff;
2-domain protein, both domains used in evaluation:
1st domain: target 2-50, 102-110 ; pdb 2-50, 102-110
2nd domain: target 51-101 ; pdb 51-101
Sequence classification:
Tandem duplicate of DUF1508 from Pfam.
Comments:
Target canceled for human predictions by organizers because of early release. Moved to the "server only" status.
Server predictions:
T0472:pdb 1-110:seq 1-110:CM_hard; alignment
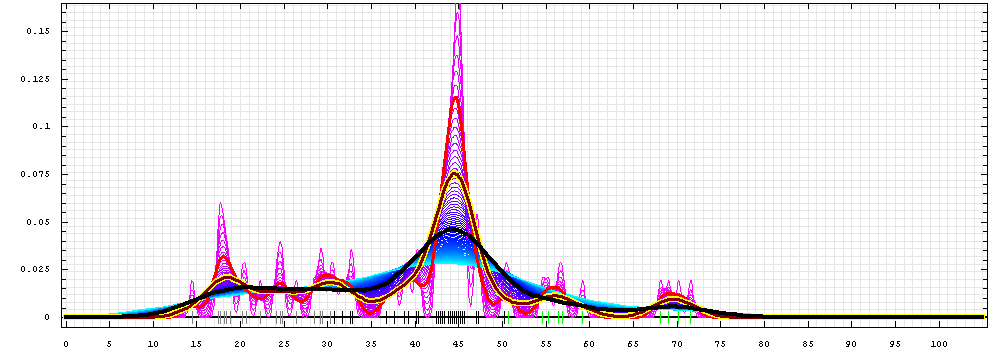
First models for T0472:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
472_1:pdb 2-50,102-110:seq 2-50,102-110:CM_easy; alignment
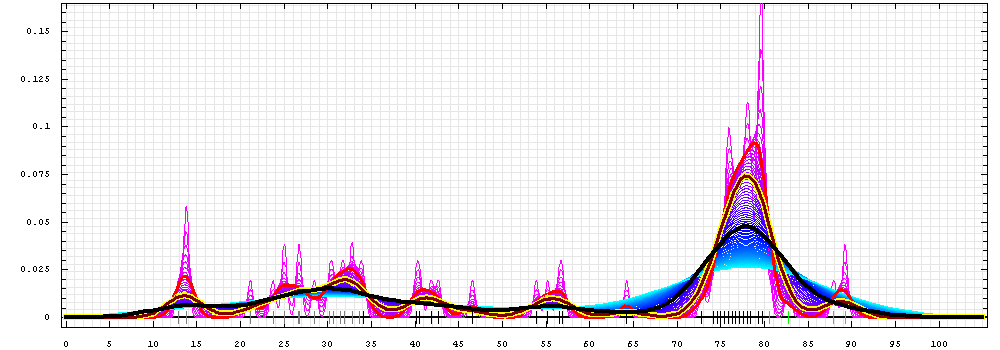
First models for T0472_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
472_2:pdb 51-101:seq 51-101:CM_easy; alignment
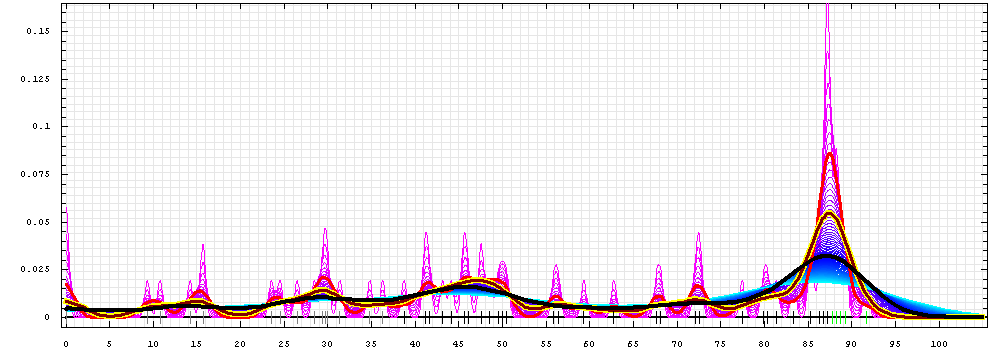
First models for T0472_2:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0472 | 472_1 | 472_2 | T0472 | 472_1 | 472_2 | T0472 | 472_1 | 472_2 | T0472 | 472_1 | 472_2 | ||||||||||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||||||||||||||
| 1 | pro−sp3−TASSER | 71.56 | 55.05 | 70.14 | 82.76 | 62.93 | 78.77 | 83.33 | 80.06 | 83.19 | 2.73 | 1.76 | 1.36 | 0.91 | 0.10 | 0.80 | 0.88 | 0.90 | 0.81 | 71.56 | 55.05 | 70.14 | 82.76 | 75.00 | 78.77 | 85.78 | 80.06 | 87.35 | 2.63 | 1.53 | 1.29 | 0.86 | 0.58 | 0.74 | 0.94 | 0.82 | 0.99 | pro−sp3−TASSER | 1 |
| 2 | FAMSD | 70.18 | 59.02 | 75.70 | 89.22 | 84.91 | 86.50 | 91.67 | 90.03 | 89.67 | 2.60 | 2.18 | 1.70 | 1.24 | 1.16 | 1.25 | 1.24 | 1.32 | 1.11 | 70.18 | 59.02 | 75.70 | 89.22 | 84.91 | 86.50 | 91.67 | 90.03 | 89.67 | 2.50 | 1.96 | 1.65 | 1.22 | 1.11 | 1.26 | 1.23 | 1.29 | 1.14 | FAMSD | 2 |
| 3 | pipe_int | 69.04 | 59.40 | 74.79 | 87.93 | 85.34 | 88.51 | 89.22 | 83.99 | 88.51 | 2.50 | 2.22 | 1.64 | 1.17 | 1.18 | 1.36 | 1.13 | 1.06 | 1.06 | 69.04 | 59.40 | 74.79 | 87.93 | 85.34 | 88.51 | 89.22 | 83.99 | 88.51 | 2.39 | 2.00 | 1.59 | 1.15 | 1.13 | 1.40 | 1.11 | 1.01 | 1.07 | pipe_int | 3 |
| 4 | RAPTOR | 68.12 | 59.71 | 68.83 | 89.22 | 86.92 | 88.91 | 80.39 | 75.49 | 73.31 | 2.41 | 2.26 | 1.28 | 1.24 | 1.26 | 1.38 | 0.75 | 0.70 | 0.35 | 68.12 | 59.71 | 68.83 | 89.22 | 86.92 | 88.91 | 80.39 | 75.49 | 73.31 | 2.30 | 2.03 | 1.20 | 1.22 | 1.21 | 1.43 | 0.67 | 0.60 | 0.11 | RAPTOR | 4 |
| 5 | FALCON_CONSENSUS | 59.17 | 46.48 | 66.48 | 77.16 | 75.14 | 73.65 | 87.25 | 85.95 | 87.33 | 1.57 | 0.85 | 1.13 | 0.63 | 0.69 | 0.50 | 1.05 | 1.15 | 1.00 | 59.17 | 53.98 | 66.72 | 78.45 | 76.44 | 74.73 | 87.75 | 85.95 | 88.33 | 1.44 | 1.42 | 1.07 | 0.62 | 0.66 | 0.47 | 1.04 | 1.10 | 1.05 | FALCON_CONSENSUS | 5 |
| 6 | FALCON | 56.88 | 53.98 | 66.72 | 78.45 | 76.44 | 74.73 | 87.75 | 85.13 | 87.99 | 1.35 | 1.65 | 1.14 | 0.69 | 0.75 | 0.57 | 1.07 | 1.11 | 1.03 | 59.17 | 53.98 | 66.72 | 78.45 | 76.72 | 75.04 | 89.22 | 87.25 | 89.76 | 1.44 | 1.42 | 1.07 | 0.62 | 0.67 | 0.49 | 1.11 | 1.16 | 1.14 | FALCON | 6 |
| 7 | PS2−server | 56.42 | 46.79 | 65.44 | 79.74 | 78.59 | 78.77 | 86.28 | 75.49 | 85.80 | 1.31 | 0.88 | 1.07 | 0.76 | 0.85 | 0.80 | 1.00 | 0.70 | 0.93 | 66.74 | 62.00 | 68.32 | 80.17 | 79.31 | 79.39 | 86.28 | 78.59 | 85.80 | 2.17 | 2.28 | 1.17 | 0.72 | 0.81 | 0.78 | 0.96 | 0.75 | 0.90 | PS2−server | 7 |
| 8 | SAM−T08−server | 55.27 | 44.80 | 61.76 | 77.59 | 73.85 | 76.75 | 67.65 | 54.25 | 70.13 | 1.20 | 0.67 | 0.84 | 0.65 | 0.63 | 0.68 | 0.20 | 0.20 | 55.96 | 50.77 | 61.76 | 78.45 | 77.87 | 77.44 | 67.65 | 62.74 | 70.13 | 1.13 | 1.07 | 0.75 | 0.62 | 0.73 | 0.65 | 0.03 | SAM−T08−server | 8 | |||
| 9 | Zhang−Server | 54.59 | 45.41 | 56.76 | 76.29 | 67.10 | 65.89 | 77.45 | 69.61 | 74.27 | 1.14 | 0.73 | 0.53 | 0.58 | 0.30 | 0.05 | 0.62 | 0.45 | 0.39 | 59.40 | 50.99 | 65.81 | 81.03 | 77.01 | 74.83 | 88.23 | 82.68 | 88.07 | 1.46 | 1.10 | 1.01 | 0.77 | 0.69 | 0.47 | 1.06 | 0.94 | 1.04 | Zhang−Server | 9 |
| 10 | METATASSER | 50.69 | 46.71 | 61.73 | 75.86 | 74.14 | 70.68 | 87.25 | 86.60 | 88.36 | 0.77 | 0.87 | 0.84 | 0.56 | 0.64 | 0.33 | 1.05 | 1.18 | 1.05 | 55.51 | 50.92 | 62.89 | 75.86 | 74.28 | 73.02 | 93.14 | 93.14 | 88.36 | 1.09 | 1.09 | 0.82 | 0.48 | 0.54 | 0.35 | 1.31 | 1.44 | 1.06 | METATASSER | 10 |
| 11 | Pcons_dot_net | 50.23 | 47.48 | 50.89 | 77.16 | 75.14 | 76.68 | 50.49 | 44.94 | 51.97 | 0.73 | 0.95 | 0.16 | 0.63 | 0.69 | 0.68 | 50.23 | 47.48 | 58.32 | 78.88 | 78.31 | 77.05 | 55.88 | 53.59 | 73.68 | 0.58 | 0.72 | 0.53 | 0.65 | 0.75 | 0.62 | 0.14 | Pcons_dot_net | 11 | |||||
| 12 | MUSTER | 47.25 | 43.73 | 63.24 | 74.14 | 72.70 | 75.78 | 87.25 | 85.95 | 87.38 | 0.45 | 0.56 | 0.93 | 0.47 | 0.57 | 0.63 | 1.05 | 1.15 | 1.00 | 47.71 | 45.72 | 64.63 | 77.59 | 75.58 | 77.25 | 88.23 | 86.60 | 89.68 | 0.34 | 0.53 | 0.93 | 0.57 | 0.61 | 0.64 | 1.06 | 1.13 | 1.14 | MUSTER | 12 |
| 13 | Phyre_de_novo | 47.02 | 41.67 | 68.47 | 77.59 | 70.11 | 78.66 | 85.29 | 81.70 | 89.82 | 0.43 | 0.34 | 1.25 | 0.65 | 0.45 | 0.79 | 0.96 | 0.97 | 1.12 | 47.02 | 45.11 | 68.47 | 77.59 | 75.72 | 78.66 | 85.29 | 81.70 | 89.82 | 0.28 | 0.47 | 1.18 | 0.57 | 0.62 | 0.73 | 0.92 | 0.90 | 1.15 | Phyre_de_novo | 13 |
| 14 | BAKER−ROBETTA | 47.02 | 39.22 | 58.32 | 76.29 | 75.72 | 74.68 | 55.88 | 53.59 | 73.68 | 0.43 | 0.08 | 0.62 | 0.58 | 0.72 | 0.56 | 0.36 | 50.23 | 47.48 | 58.32 | 78.88 | 78.31 | 77.05 | 55.88 | 53.59 | 73.68 | 0.58 | 0.72 | 0.53 | 0.65 | 0.75 | 0.62 | 0.14 | BAKER−ROBETTA | 14 | ||||
| 15 | fais−server | 45.64 | 43.81 | 66.76 | 79.74 | 79.74 | 77.48 | 89.22 | 84.64 | 89.77 | 0.30 | 0.56 | 1.15 | 0.76 | 0.91 | 0.72 | 1.13 | 1.09 | 1.12 | 48.85 | 46.94 | 67.89 | 84.48 | 84.48 | 80.47 | 91.18 | 89.54 | 89.79 | 0.45 | 0.66 | 1.14 | 0.96 | 1.08 | 0.85 | 1.21 | 1.27 | 1.15 | fais−server | 15 |
| 16 | GS−KudlatyPred | 45.64 | 43.35 | 61.16 | 78.45 | 76.72 | 77.73 | 79.90 | 78.92 | 78.52 | 0.30 | 0.52 | 0.80 | 0.69 | 0.77 | 0.74 | 0.73 | 0.85 | 0.59 | 45.64 | 43.35 | 61.16 | 78.45 | 76.72 | 77.73 | 79.90 | 78.92 | 78.52 | 0.14 | 0.28 | 0.71 | 0.62 | 0.67 | 0.67 | 0.65 | 0.76 | 0.44 | GS−KudlatyPred | 16 |
| 17 | FOLDpro | 45.41 | 43.42 | 46.69 | 80.60 | 77.73 | 77.53 | 31.37 | 25.98 | 39.62 | 0.27 | 0.52 | 0.80 | 0.81 | 0.73 | 45.41 | 43.42 | 46.69 | 80.60 | 77.73 | 77.53 | 48.04 | 46.57 | 60.22 | 0.12 | 0.28 | 0.74 | 0.72 | 0.65 | FOLDpro | 17 | ||||||||
| 18 | Phragment | 45.41 | 42.81 | 57.12 | 75.43 | 71.69 | 76.65 | 72.06 | 71.41 | 71.92 | 0.27 | 0.46 | 0.55 | 0.54 | 0.52 | 0.68 | 0.39 | 0.53 | 0.28 | 47.71 | 44.42 | 57.12 | 76.72 | 74.14 | 77.27 | 72.55 | 71.89 | 71.92 | 0.34 | 0.39 | 0.45 | 0.53 | 0.53 | 0.64 | 0.28 | 0.43 | 0.03 | Phragment | 18 |
| 19 | MULTICOM−CLUSTER | 45.18 | 39.68 | 64.56 | 79.74 | 78.31 | 78.13 | 88.72 | 86.77 | 86.12 | 0.25 | 0.12 | 1.01 | 0.76 | 0.84 | 0.76 | 1.11 | 1.18 | 0.94 | 46.79 | 44.49 | 67.43 | 79.74 | 78.31 | 78.68 | 89.22 | 87.25 | 88.28 | 0.25 | 0.40 | 1.11 | 0.69 | 0.75 | 0.73 | 1.11 | 1.16 | 1.05 | MULTICOM−CLUSTER | 19 |
| 20 | HHpred4 | 44.95 | 43.88 | 64.26 | 79.31 | 77.59 | 77.04 | 87.25 | 87.25 | 88.45 | 0.23 | 0.57 | 0.99 | 0.73 | 0.81 | 0.70 | 1.05 | 1.20 | 1.05 | 44.95 | 43.88 | 64.26 | 79.31 | 77.59 | 77.04 | 87.25 | 87.25 | 88.45 | 0.08 | 0.33 | 0.91 | 0.67 | 0.72 | 0.62 | 1.01 | 1.16 | 1.06 | HHpred4 | 20 |
| 21 | HHpred2 | 44.95 | 43.88 | 64.26 | 79.31 | 77.59 | 77.04 | 87.25 | 87.25 | 88.45 | 0.23 | 0.57 | 0.99 | 0.73 | 0.81 | 0.70 | 1.05 | 1.20 | 1.05 | 44.95 | 43.88 | 64.26 | 79.31 | 77.59 | 77.04 | 87.25 | 87.25 | 88.45 | 0.08 | 0.33 | 0.91 | 0.67 | 0.72 | 0.62 | 1.01 | 1.16 | 1.06 | HHpred2 | 21 |
| 22 | HHpred5 | 44.95 | 43.88 | 64.26 | 79.31 | 77.59 | 77.04 | 87.25 | 87.25 | 88.45 | 0.23 | 0.57 | 0.99 | 0.73 | 0.81 | 0.70 | 1.05 | 1.20 | 1.05 | 44.95 | 43.88 | 64.26 | 79.31 | 77.59 | 77.04 | 87.25 | 87.25 | 88.45 | 0.08 | 0.33 | 0.91 | 0.67 | 0.72 | 0.62 | 1.01 | 1.16 | 1.06 | HHpred5 | 22 |
| 23 | MULTICOM−REFINE | 44.95 | 43.73 | 64.41 | 79.74 | 78.31 | 77.92 | 88.23 | 87.25 | 86.17 | 0.23 | 0.56 | 1.00 | 0.76 | 0.84 | 0.75 | 1.09 | 1.20 | 0.95 | 47.48 | 44.49 | 67.14 | 79.74 | 78.31 | 78.09 | 89.22 | 87.91 | 88.25 | 0.32 | 0.40 | 1.09 | 0.69 | 0.75 | 0.69 | 1.11 | 1.19 | 1.05 | MULTICOM−REFINE | 23 |
| 24 | MUProt | 44.95 | 43.12 | 64.30 | 79.74 | 78.31 | 77.69 | 88.23 | 87.91 | 86.19 | 0.23 | 0.49 | 0.99 | 0.76 | 0.84 | 0.74 | 1.09 | 1.23 | 0.95 | 47.94 | 43.58 | 66.93 | 79.74 | 78.31 | 77.83 | 89.71 | 87.91 | 88.00 | 0.37 | 0.30 | 1.08 | 0.69 | 0.75 | 0.68 | 1.14 | 1.19 | 1.03 | MUProt | 24 |
| 25 | BioSerf | 44.95 | 42.20 | 54.20 | 79.74 | 63.36 | 77.83 | 44.12 | 41.99 | 59.15 | 0.23 | 0.39 | 0.37 | 0.76 | 0.12 | 0.74 | 44.95 | 42.20 | 54.20 | 79.74 | 63.36 | 77.83 | 44.12 | 41.99 | 59.15 | 0.08 | 0.15 | 0.26 | 0.69 | 0.68 | BioSerf | 25 | |||||||
| 26 | MULTICOM−CMFR | 44.73 | 43.65 | 64.60 | 79.74 | 78.59 | 78.12 | 88.23 | 87.91 | 86.42 | 0.21 | 0.55 | 1.01 | 0.76 | 0.85 | 0.76 | 1.09 | 1.23 | 0.96 | 47.02 | 43.81 | 67.16 | 79.74 | 78.59 | 78.21 | 90.20 | 88.89 | 88.12 | 0.28 | 0.33 | 1.10 | 0.69 | 0.77 | 0.70 | 1.16 | 1.24 | 1.04 | MULTICOM−CMFR | 26 |
| 27 | GS−MetaServer2 | 44.73 | 43.65 | 26.99 | 13.79 | 13.22 | 4.72 | 86.77 | 86.77 | 87.61 | 0.21 | 0.55 | 1.03 | 1.18 | 1.01 | 44.73 | 43.65 | 35.45 | 33.62 | 27.01 | 34.94 | 86.77 | 86.77 | 87.61 | 0.06 | 0.31 | 0.99 | 1.14 | 1.01 | GS−MetaServer2 | 27 | ||||||||
| 28 | FFASflextemplate | 44.73 | 43.35 | 26.51 | 13.79 | 12.93 | 4.65 | 86.77 | 86.11 | 87.08 | 0.21 | 0.52 | 1.03 | 1.15 | 0.99 | 44.73 | 43.35 | 26.51 | 13.79 | 12.93 | 4.65 | 86.77 | 86.11 | 87.08 | 0.06 | 0.28 | 0.99 | 1.11 | 0.98 | FFASflextemplate | 28 | ||||||||
| 29 | MULTICOM−RANK | 44.49 | 43.88 | 64.02 | 79.31 | 77.87 | 77.22 | 88.23 | 87.91 | 86.07 | 0.19 | 0.57 | 0.98 | 0.73 | 0.82 | 0.71 | 1.09 | 1.23 | 0.94 | 47.71 | 43.88 | 66.51 | 79.31 | 77.87 | 77.49 | 89.71 | 87.91 | 87.79 | 0.34 | 0.33 | 1.05 | 0.67 | 0.73 | 0.65 | 1.14 | 1.19 | 1.02 | MULTICOM−RANK | 29 |
| 30 | Poing | 44.27 | 40.29 | 57.75 | 76.72 | 73.85 | 77.30 | 72.55 | 70.59 | 72.16 | 0.17 | 0.19 | 0.59 | 0.60 | 0.63 | 0.71 | 0.41 | 0.50 | 0.29 | 45.87 | 41.90 | 57.75 | 76.72 | 73.85 | 77.53 | 72.55 | 70.59 | 72.16 | 0.17 | 0.12 | 0.49 | 0.53 | 0.52 | 0.65 | 0.28 | 0.37 | 0.04 | Poing | 30 |
| 31 | Phyre2 | 44.27 | 40.29 | 57.74 | 76.72 | 73.85 | 77.42 | 72.55 | 68.95 | 71.96 | 0.17 | 0.19 | 0.59 | 0.60 | 0.63 | 0.72 | 0.41 | 0.43 | 0.28 | 45.87 | 44.04 | 57.74 | 76.72 | 74.14 | 77.42 | 73.53 | 68.95 | 71.96 | 0.17 | 0.35 | 0.49 | 0.53 | 0.53 | 0.65 | 0.33 | 0.29 | 0.03 | Phyre2 | 31 |
| 32 | nFOLD3 | 44.04 | 40.98 | 27.55 | 75.86 | 75.29 | 70.92 | 15.69 | 14.71 | 3.34 | 0.14 | 0.26 | 0.56 | 0.70 | 0.35 | 44.04 | 42.66 | 40.10 | 79.31 | 78.16 | 77.25 | 65.69 | 58.50 | 71.83 | 0.20 | 0.67 | 0.75 | 0.64 | 0.02 | nFOLD3 | 32 | ||||||||
| 33 | FUGUE_KM | 44.04 | 40.98 | 27.55 | 75.86 | 75.29 | 70.92 | 15.69 | 14.71 | 3.34 | 0.14 | 0.26 | 0.56 | 0.70 | 0.35 | 44.04 | 40.98 | 27.55 | 75.86 | 75.29 | 70.92 | 51.47 | 42.97 | 58.16 | 0.02 | 0.48 | 0.59 | 0.21 | FUGUE_KM | 33 | |||||||||
| 34 | COMA | 43.81 | 39.30 | 64.87 | 78.02 | 69.68 | 77.55 | 87.25 | 74.84 | 87.46 | 0.12 | 0.08 | 1.03 | 0.67 | 0.43 | 0.73 | 1.05 | 0.68 | 1.01 | 43.81 | 39.30 | 64.87 | 78.02 | 69.68 | 77.55 | 87.25 | 74.84 | 87.46 | 0.95 | 0.60 | 0.30 | 0.66 | 1.01 | 0.57 | 1.00 | COMA | 34 | ||
| 35 | COMA−M | 43.81 | 39.30 | 64.87 | 78.02 | 69.68 | 77.55 | 87.25 | 74.84 | 87.46 | 0.12 | 0.08 | 1.03 | 0.67 | 0.43 | 0.73 | 1.05 | 0.68 | 1.01 | 43.81 | 39.30 | 64.87 | 78.02 | 69.68 | 77.55 | 87.25 | 74.84 | 87.46 | 0.95 | 0.60 | 0.30 | 0.66 | 1.01 | 0.57 | 1.00 | COMA−M | 35 | ||
| 36 | FFASstandard | 43.35 | 39.22 | 26.26 | 13.79 | 12.93 | 4.57 | 85.29 | 79.74 | 86.57 | 0.08 | 0.08 | 0.96 | 0.88 | 0.97 | 44.73 | 43.35 | 26.51 | 13.79 | 12.93 | 4.65 | 86.77 | 86.11 | 87.08 | 0.06 | 0.28 | 0.99 | 1.11 | 0.98 | FFASstandard | 36 | ||||||||
| 37 | panther_server | 43.35 | 38.61 | 28.45 | 78.02 | 76.29 | 71.54 | 9.31 | 8.66 | 1.90 | 0.08 | 0.01 | 0.67 | 0.74 | 0.38 | 43.35 | 38.61 | 28.45 | 78.02 | 76.29 | 71.54 | 25.49 | 20.26 | 30.22 | 0.60 | 0.65 | 0.25 | panther_server | 37 | ||||||||||
| 38 | rehtnap | 43.12 | 38.69 | 27.04 | 75.00 | 73.85 | 70.71 | 10.78 | 10.78 | 2.00 | 0.06 | 0.02 | 0.52 | 0.63 | 0.33 | 43.58 | 42.35 | 27.53 | 75.86 | 73.85 | 70.71 | 17.16 | 15.85 | 5.16 | 0.17 | 0.48 | 0.52 | 0.19 | rehtnap | 38 | |||||||||
| 39 | CpHModels | 42.89 | 41.82 | 27.72 | 74.57 | 73.13 | 70.82 | 14.22 | 13.89 | 3.10 | 0.04 | 0.35 | 0.49 | 0.59 | 0.34 | 42.89 | 41.82 | 27.72 | 74.57 | 73.13 | 70.82 | 14.22 | 13.89 | 3.10 | 0.11 | 0.40 | 0.48 | 0.20 | CpHModels | 39 | |||||||||
| 40 | 3Dpro | 42.66 | 40.21 | 52.64 | 78.45 | 76.72 | 77.49 | 45.59 | 42.32 | 56.59 | 0.02 | 0.18 | 0.27 | 0.69 | 0.77 | 0.72 | 46.79 | 43.58 | 54.84 | 80.60 | 77.59 | 77.76 | 50.00 | 45.91 | 63.47 | 0.25 | 0.30 | 0.30 | 0.74 | 0.72 | 0.67 | 3Dpro | 40 | ||||||
| 41 | Pushchino | 42.43 | 40.44 | 25.87 | 78.02 | 74.86 | 70.40 | 0.00 | 0.00 | 0.00 | 0.21 | 0.67 | 0.68 | 0.32 | 42.43 | 40.44 | 25.87 | 78.02 | 74.86 | 70.40 | 0.00 | 0.00 | 0.00 | 0.60 | 0.57 | 0.17 | Pushchino | 41 | |||||||||||
| 42 | SAM−T02−server | 40.37 | 39.76 | 25.33 | 75.86 | 75.29 | 70.92 | 0.00 | 0.00 | 0.00 | 0.13 | 0.56 | 0.70 | 0.35 | 40.37 | 39.76 | 25.33 | 75.86 | 75.29 | 70.92 | 44.61 | 38.07 | 53.62 | 0.48 | 0.59 | 0.21 | SAM−T02−server | 42 | |||||||||||
| 43 | Pcons_local | 40.14 | 38.00 | 25.05 | 12.93 | 12.93 | 4.65 | 81.37 | 76.80 | 83.10 | 0.79 | 0.76 | 0.80 | 40.14 | 38.00 | 27.05 | 43.10 | 42.24 | 45.19 | 81.37 | 76.80 | 83.10 | 0.72 | 0.66 | 0.73 | Pcons_local | 43 | ||||||||||||
| 44 | Distill | 39.22 | 35.55 | 47.62 | 64.22 | 58.76 | 61.82 | 41.18 | 34.31 | 50.60 | 39.22 | 35.55 | 47.62 | 64.22 | 58.76 | 63.58 | 41.18 | 34.31 | 50.60 | Distill | 44 | ||||||||||||||||||
| 45 | Fiser−M4T | 38.76 | 38.30 | 24.38 | 72.84 | 72.27 | 68.65 | 0.00 | 0.00 | 0.00 | 0.41 | 0.55 | 0.21 | 38.76 | 38.30 | 24.38 | 72.84 | 72.27 | 68.65 | 0.00 | 0.00 | 0.00 | 0.31 | 0.43 | 0.05 | Fiser−M4T | 45 | ||||||||||||
| 46 | SAM−T06−server | 37.62 | 29.89 | 50.83 | 55.17 | 52.30 | 65.68 | 48.53 | 41.99 | 62.72 | 0.16 | 0.04 | 40.37 | 39.60 | 50.83 | 74.14 | 73.56 | 68.35 | 48.53 | 41.99 | 62.72 | 0.04 | 0.38 | 0.50 | 0.03 | SAM−T06−server | 46 | ||||||||||||
| 47 | GeneSilicoMetaServer | 36.70 | 32.11 | 35.45 | 25.00 | 22.13 | 27.79 | 68.14 | 61.27 | 71.18 | 0.22 | 0.10 | 0.25 | 44.73 | 43.65 | 35.45 | 33.62 | 27.01 | 34.94 | 86.77 | 86.77 | 87.61 | 0.06 | 0.31 | 0.99 | 1.14 | 1.01 | GeneSilicoMetaServer | 47 | ||||||||||
| 48 | Frankenstein | 32.80 | 26.07 | 35.60 | 33.62 | 25.29 | 33.96 | 62.74 | 50.00 | 65.74 | 34.86 | 29.36 | 39.54 | 38.79 | 35.92 | 47.89 | 62.74 | 54.58 | 65.91 | Frankenstein | 48 | ||||||||||||||||||
| 49 | 3DShot2 | 32.57 | 26.91 | 28.35 | 31.90 | 25.00 | 28.69 | 56.37 | 49.18 | 56.29 | 32.57 | 26.91 | 28.35 | 31.90 | 25.00 | 28.69 | 56.37 | 49.18 | 56.29 | 3DShot2 | 49 | ||||||||||||||||||
| 50 | RBO−Proteus | 31.65 | 27.68 | 44.88 | 56.90 | 50.86 | 64.54 | 59.31 | 46.57 | 53.15 | 31.65 | 28.21 | 44.88 | 59.05 | 53.30 | 64.54 | 59.31 | 50.33 | 55.30 | RBO−Proteus | 50 | ||||||||||||||||||
| 51 | FFASsuboptimal | 30.73 | 29.51 | 32.56 | 42.67 | 36.06 | 45.88 | 49.51 | 40.03 | 45.42 | 43.35 | 42.58 | 33.57 | 78.88 | 78.31 | 74.66 | 49.51 | 40.03 | 45.42 | 0.19 | 0.65 | 0.75 | 0.46 | FFASsuboptimal | 51 | ||||||||||||||
| 52 | PSI | 30.27 | 27.22 | 47.36 | 56.47 | 51.29 | 59.79 | 47.55 | 39.87 | 65.77 | 32.57 | 30.12 | 48.25 | 60.34 | 57.18 | 61.38 | 48.04 | 42.16 | 66.05 | PSI | 52 | ||||||||||||||||||
| 53 | MUFOLD−Server | 29.36 | 24.31 | 32.12 | 53.88 | 44.54 | 46.65 | 34.80 | 33.17 | 43.18 | 29.59 | 24.31 | 39.38 | 54.31 | 44.54 | 46.65 | 49.51 | 43.46 | 61.79 | MUFOLD−Server | 53 | ||||||||||||||||||
| 54 | FEIG | 29.13 | 24.39 | 34.54 | 40.52 | 33.91 | 48.04 | 41.67 | 39.71 | 46.62 | 29.13 | 26.84 | 38.36 | 42.67 | 36.49 | 52.96 | 47.06 | 42.48 | 50.35 | FEIG | 54 | ||||||||||||||||||
| 55 | MUFOLD−MD | 28.44 | 23.70 | 48.78 | 46.55 | 42.82 | 61.46 | 50.00 | 41.18 | 67.43 | 0.03 | 0.07 | 37.62 | 34.86 | 55.66 | 63.36 | 60.34 | 77.09 | 50.00 | 43.46 | 67.43 | 0.35 | 0.63 | MUFOLD−MD | 55 | ||||||||||||||
| 56 | mGenTHREADER | 26.38 | 24.62 | 44.90 | 40.09 | 38.65 | 71.33 | 47.55 | 41.99 | 44.98 | 0.37 | 26.38 | 24.62 | 44.90 | 40.09 | 38.65 | 71.33 | 47.55 | 41.99 | 44.98 | 0.23 | mGenTHREADER | 56 | ||||||||||||||||
| 57 | circle | 24.77 | 21.86 | 30.32 | 41.81 | 36.64 | 41.41 | 43.14 | 42.48 | 47.06 | 30.50 | 25.61 | 37.01 | 54.31 | 44.54 | 52.31 | 48.04 | 46.41 | 49.20 | circle | 57 | ||||||||||||||||||
| 58 | 3D−JIGSAW_AEP | 24.54 | 22.94 | 33.76 | 32.76 | 32.47 | 34.64 | 46.08 | 41.18 | 53.27 | 24.54 | 22.94 | 34.14 | 33.19 | 32.47 | 35.44 | 46.08 | 41.18 | 53.49 | 3D−JIGSAW_AEP | 58 | ||||||||||||||||||
| 59 | 3D−JIGSAW_V3 | 24.08 | 23.01 | 36.31 | 34.05 | 27.73 | 42.16 | 45.59 | 42.97 | 52.23 | 25.69 | 23.01 | 41.85 | 37.93 | 33.91 | 51.86 | 46.57 | 42.97 | 54.58 | 3D−JIGSAW_V3 | 59 | ||||||||||||||||||
| 60 | forecast | 22.25 | 20.26 | 31.37 | 32.76 | 24.71 | 37.48 | 41.18 | 37.26 | 51.56 | 23.62 | 20.72 | 33.15 | 32.76 | 25.43 | 37.48 | 44.12 | 41.83 | 59.25 | forecast | 60 | ||||||||||||||||||
| 61 | LOOPP_Server | 20.64 | 16.51 | 24.16 | 30.17 | 23.42 | 29.69 | 36.27 | 29.09 | 43.14 | 35.32 | 29.97 | 32.78 | 38.36 | 32.62 | 38.72 | 64.22 | 55.72 | 64.08 | LOOPP_Server | 61 | ||||||||||||||||||
| 62 | mariner1 | 20.18 | 15.44 | 23.02 | 31.47 | 25.72 | 36.37 | 28.43 | 23.86 | 30.15 | 21.79 | 19.95 | 29.04 | 35.34 | 31.32 | 45.89 | 39.71 | 37.58 | 44.66 | mariner1 | 62 | ||||||||||||||||||
| 63 | keasar−server | 18.81 | 16.21 | 29.13 | 28.45 | 22.13 | 34.20 | 38.73 | 33.17 | 49.66 | 27.75 | 24.85 | 31.25 | 40.52 | 35.06 | 36.34 | 52.45 | 43.79 | 55.94 | keasar−server | 63 | ||||||||||||||||||
| 64 | ACOMPMOD | 18.35 | 15.67 | 23.45 | 25.00 | 22.70 | 28.43 | 29.41 | 25.98 | 44.62 | 20.64 | 18.73 | 23.45 | 31.90 | 28.74 | 38.44 | 30.39 | 26.14 | 44.62 | ACOMPMOD | 64 | ||||||||||||||||||
| 65 | huber−torda−server | 18.12 | 13.69 | 15.42 | 23.71 | 18.82 | 22.83 | 29.90 | 27.61 | 31.17 | 18.12 | 17.05 | 21.83 | 32.76 | 28.74 | 31.50 | 29.90 | 29.57 | 35.45 | huber−torda−server | 65 | ||||||||||||||||||
| 66 | RANDOM | 17.70 | 14.78 | 17.70 | 26.68 | 22.06 | 26.68 | 29.71 | 25.56 | 29.71 | 17.70 | 14.78 | 17.70 | 26.68 | 22.06 | 26.68 | 29.71 | 25.56 | 29.71 | RANDOM | 66 | ||||||||||||||||||
| 67 | schenk−torda−server | 17.66 | 13.23 | 16.09 | 26.72 | 22.13 | 26.69 | 26.47 | 25.16 | 28.15 | 18.35 | 17.28 | 22.31 | 30.17 | 29.89 | 35.02 | 29.90 | 26.47 | 41.87 | schenk−torda−server | 67 | ||||||||||||||||||
| 68 | Pcons_multi | 17.43 | 15.06 | 20.58 | 30.60 | 26.58 | 37.25 | 24.51 | 22.55 | 24.39 | 21.33 | 16.97 | 24.91 | 30.60 | 26.87 | 37.25 | 33.82 | 28.43 | 45.17 | Pcons_multi | 68 | ||||||||||||||||||
| 69 | OLGAFS | 14.45 | 11.54 | 13.95 | 21.12 | 16.24 | 23.18 | 23.53 | 21.57 | 26.55 | 17.20 | 16.59 | 19.28 | 23.71 | 18.82 | 24.05 | 32.35 | 31.54 | 39.63 | OLGAFS | 69 | ||||||||||||||||||
| 70 | BHAGEERATH | BHAGEERATH | 70 | ||||||||||||||||||||||||||||||||||||
| 71 | LEE−SERVER | LEE−SERVER | 71 | ||||||||||||||||||||||||||||||||||||
| 72 | YASARA | YASARA | 72 | ||||||||||||||||||||||||||||||||||||
| 73 | mahmood−torda−server | mahmood−torda−server | 73 | ||||||||||||||||||||||||||||||||||||
| 74 | test_http_server_01 | test_http_server_01 | 74 | ||||||||||||||||||||||||||||||||||||