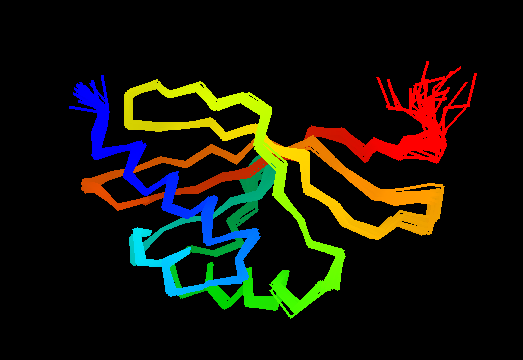
T0475
protein Atu0742 from Agrobacterium tumefaciens
Target sequence:
>T0475 AtT8, unknown, 122 residues
LNSEIELPVQKQLEAYNARDIDAFMAWWADDCQYYAFPATLLAGNAAEIRVRHIERFKEPDLYGELLTRVIVGNVVIDHETVTRNFPEGKGEVDVACIYEVENGRIAKAWFKIGEPRIVSQK
Structure:
Method: NMR
Determined by: NESG
PDB ID: 2k54
Domains: PyMOL of domains
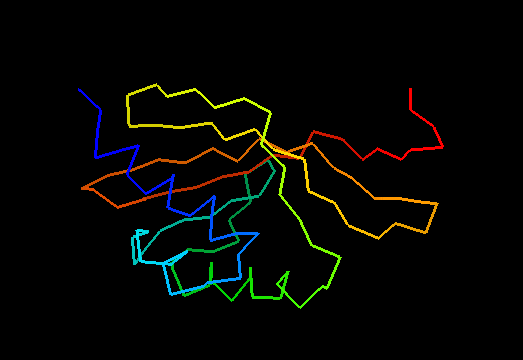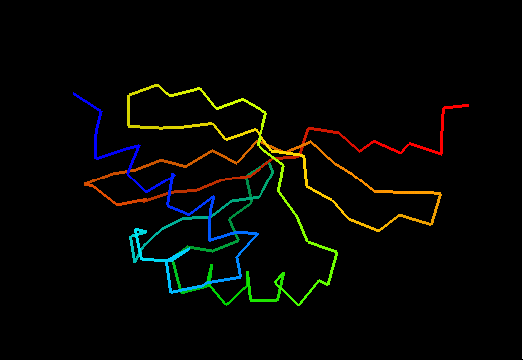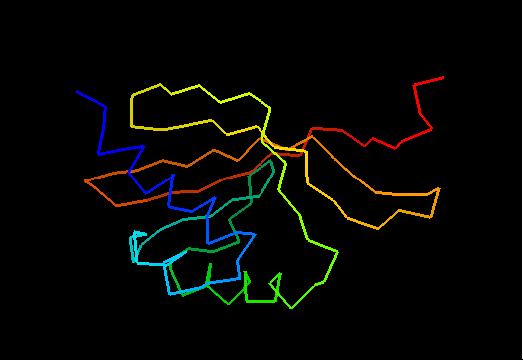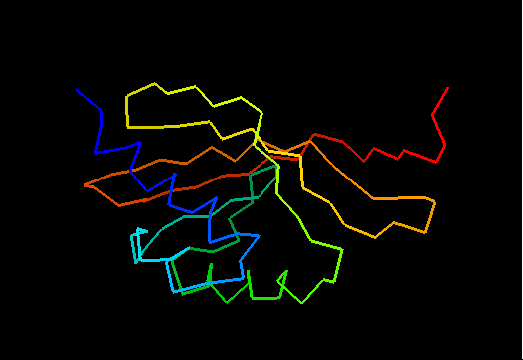
Single domain protein. Good NMR structure, only the last residue poorly ordered, but it is absent in the target sequence and was removed anyway.
Structure classification:
Cystatin-like fold, similar to Nuclear transport factor 2 (NTF2-like) proteins.
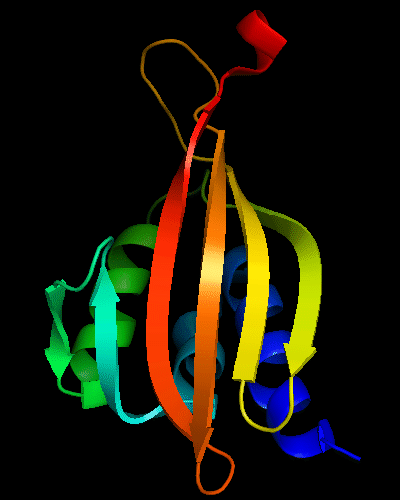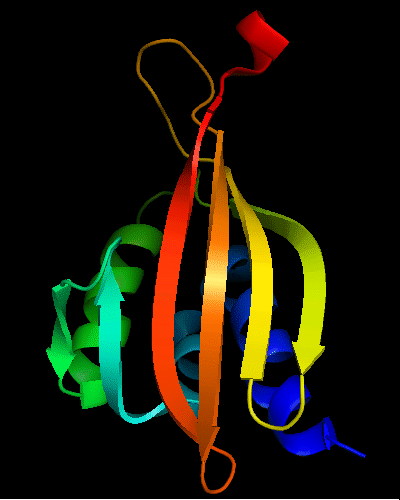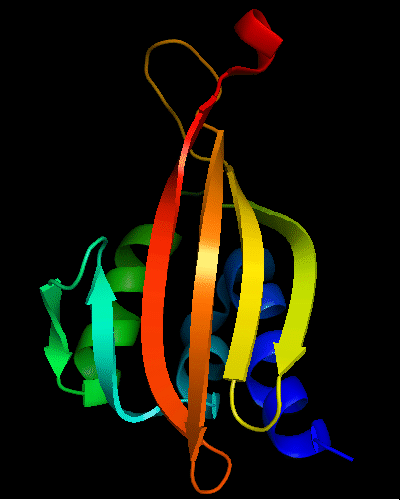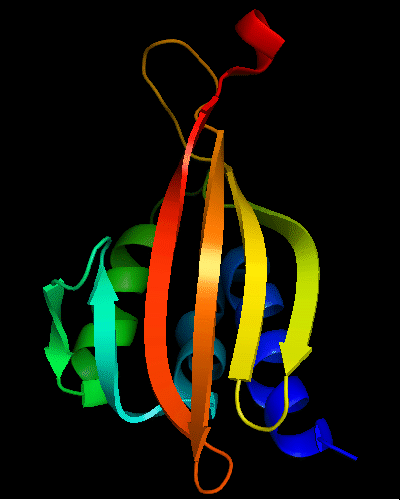
Cartoon diagram of T0475: 2k54 model 1 residues 1-122
CASP category:
Comparative modeling: medium. Similar to the templates. Differences are in loops and around the C-terminus, which has a helical turn in templates, but is extended in the target NMR structure.
Closest templates:
Ketosteroid isomerase, e.g. 1isk.
Target sequence - PDB file inconsistencies:
The first residue is Leu in the target, but Met in PDB. The last PDB residue Ser123 is not in the target sequence, but it is not ordered in NMR models anyway.
T0475 2k54.pdb T0475.pdb PyMOL PyMOL of domains
T0475 1 LNSEIELPVQKQLEAYNARDIDAFMAWWADDCQYYAFPATLLAGNAAEIRVRHIERFKEPDLYGELLTRVIVGNVVIDHETVTRNFPEGKGEVDVACIYEVENGRIAKAWFKIGEPRIVSQK- 122 :|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~ 2k54A 1 MNSEIELPVQKQLEAYNARDIDAFMAWWADDCQYYAFPATLLAGNAAEIRVRHIERFKEPDLYGELLTRVIVGNVVIDHETVTRNFPEGKGEVDVACIYEVENGRIAKAWFKIGEPRIVSQKS 123
Residue change log: change 1 M to L; remove 123 S as it is not present in target sequence;
Single domain protein: target 1-122 ; pdb 1-122
Sequence classification:
SnoaL-like polyketide cyclase family (SnoaL) in Pfam.
Comments:
Server predictions:
T0475:pdb 1-122:seq 1-122:CM_medium; alignment
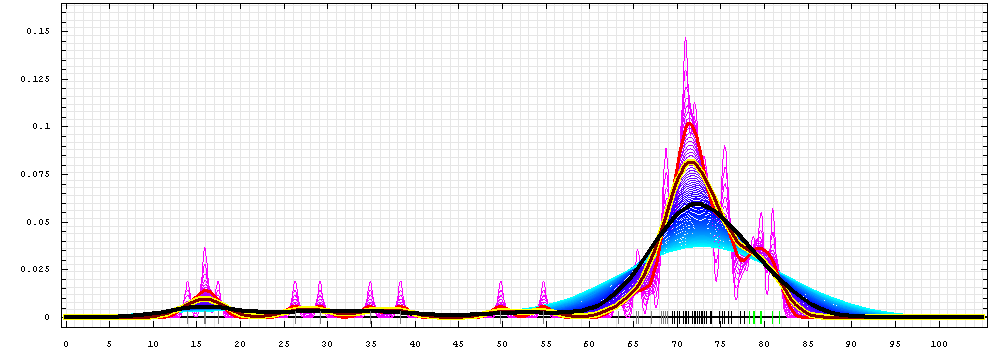
First models for T0475:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0475 | T0475 | T0475 | T0475 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | HHpred5 | 81.76 | 72.34 | 80.18 | 1.76 | 1.28 | 1.16 | 81.76 | 72.34 | 80.18 | 1.75 | 0.92 | 1.27 | HHpred5 | 1 |
| 2 | MULTICOM−CMFR | 80.94 | 77.12 | 78.42 | 1.60 | 2.07 | 0.91 | 80.94 | 77.12 | 78.42 | 1.55 | 1.95 | 0.87 | MULTICOM−CMFR | 2 |
| 3 | MUProt | 80.94 | 75.75 | 76.74 | 1.60 | 1.85 | 0.68 | 81.76 | 76.43 | 77.58 | 1.75 | 1.80 | 0.68 | MUProt | 3 |
| 4 | MULTICOM−REFINE | 80.94 | 71.24 | 78.14 | 1.60 | 1.10 | 0.88 | 83.20 | 76.91 | 78.86 | 2.12 | 1.91 | 0.97 | MULTICOM−REFINE | 4 |
| 5 | Phyre_de_novo | 79.71 | 75.75 | 79.52 | 1.38 | 1.85 | 1.07 | 79.71 | 75.75 | 79.52 | 1.24 | 1.66 | 1.12 | Phyre_de_novo | 5 |
| 6 | LEE−SERVER | 79.71 | 74.80 | 77.96 | 1.38 | 1.69 | 0.85 | 80.94 | 76.03 | 78.39 | 1.55 | 1.72 | 0.86 | LEE−SERVER | 6 |
| 7 | Zhang−Server | 79.51 | 75.96 | 81.19 | 1.34 | 1.88 | 1.30 | 79.51 | 75.96 | 81.19 | 1.19 | 1.70 | 1.49 | Zhang−Server | 7 |
| 8 | METATASSER | 78.89 | 67.28 | 77.21 | 1.23 | 0.44 | 0.75 | 79.30 | 74.93 | 79.22 | 1.14 | 1.48 | 1.05 | METATASSER | 8 |
| 9 | GS−KudlatyPred | 78.69 | 73.50 | 76.69 | 1.19 | 1.47 | 0.67 | 78.69 | 73.50 | 76.69 | 0.98 | 1.17 | 0.48 | GS−KudlatyPred | 9 |
| 10 | COMA−M | 78.28 | 68.99 | 77.00 | 1.11 | 0.72 | 0.72 | 79.92 | 75.00 | 77.84 | 1.29 | 1.49 | 0.74 | COMA−M | 10 |
| 11 | GeneSilicoMetaServer | 77.66 | 70.97 | 75.72 | 1.00 | 1.05 | 0.54 | 77.66 | 70.97 | 75.72 | 0.73 | 0.62 | 0.26 | GeneSilicoMetaServer | 11 |
| 12 | PS2−server | 77.25 | 72.61 | 74.89 | 0.92 | 1.32 | 0.42 | 77.25 | 72.61 | 74.89 | 0.62 | 0.98 | 0.07 | PS2−server | 12 |
| 13 | pro−sp3−TASSER | 76.23 | 65.16 | 80.00 | 0.74 | 0.09 | 1.13 | 79.30 | 74.93 | 80.38 | 1.14 | 1.48 | 1.31 | pro−sp3−TASSER | 13 |
| 14 | FFASstandard | 75.82 | 65.03 | 72.09 | 0.66 | 0.06 | 0.03 | 76.43 | 69.54 | 75.70 | 0.42 | 0.32 | 0.25 | FFASstandard | 14 |
| 15 | FFASsuboptimal | 75.82 | 60.52 | 72.16 | 0.66 | 0.04 | 76.03 | 69.94 | 72.26 | 0.32 | 0.40 | FFASsuboptimal | 15 | ||
| 16 | LOOPP_Server | 75.41 | 72.68 | 71.50 | 0.59 | 1.34 | 77.25 | 72.68 | 74.16 | 0.62 | 0.99 | LOOPP_Server | 16 | ||
| 17 | 3DShot2 | 75.41 | 66.12 | 73.98 | 0.59 | 0.25 | 0.30 | 75.41 | 66.12 | 73.98 | 0.16 | 3DShot2 | 17 | ||
| 18 | Frankenstein | 75.41 | 62.43 | 70.11 | 0.59 | 75.41 | 63.93 | 76.00 | 0.16 | 0.32 | Frankenstein | 18 | |||
| 19 | MUSTER | 75.20 | 70.56 | 73.83 | 0.55 | 0.98 | 0.28 | 79.71 | 73.57 | 77.63 | 1.24 | 1.19 | 0.69 | MUSTER | 19 |
| 20 | circle | 74.80 | 66.60 | 72.40 | 0.47 | 0.33 | 0.08 | 75.61 | 66.60 | 72.72 | 0.21 | circle | 20 | ||
| 21 | fais−server | 73.97 | 67.69 | 78.73 | 0.32 | 0.51 | 0.96 | 73.97 | 67.69 | 78.73 | 0.94 | fais−server | 21 | ||
| 22 | Pcons_dot_net | 73.77 | 63.32 | 69.19 | 0.28 | 77.05 | 67.01 | 72.88 | 0.57 | Pcons_dot_net | 22 | ||||
| 23 | Pcons_multi | 73.77 | 63.32 | 69.19 | 0.28 | 77.46 | 70.36 | 75.24 | 0.68 | 0.49 | 0.15 | Pcons_multi | 23 | ||
| 24 | MULTICOM−CLUSTER | 73.36 | 69.13 | 67.32 | 0.21 | 0.75 | 77.66 | 72.20 | 74.58 | 0.73 | 0.89 | MULTICOM−CLUSTER | 24 | ||
| 25 | 3D−JIGSAW_AEP | 73.16 | 65.64 | 77.66 | 0.17 | 0.17 | 0.81 | 73.16 | 65.64 | 77.66 | 0.70 | 3D−JIGSAW_AEP | 25 | ||
| 26 | SAM−T08−server | 73.16 | 63.66 | 75.44 | 0.17 | 0.50 | 73.97 | 67.01 | 75.44 | 0.19 | SAM−T08−server | 26 | |||
| 27 | panther_server | 72.95 | 66.67 | 77.10 | 0.13 | 0.34 | 0.73 | 72.95 | 66.67 | 77.10 | 0.57 | panther_server | 27 | ||
| 28 | RAPTOR | 72.75 | 66.19 | 73.80 | 0.10 | 0.26 | 0.27 | 73.16 | 67.69 | 75.27 | 0.15 | RAPTOR | 28 | ||
| 29 | MUFOLD−Server | 72.54 | 65.85 | 75.91 | 0.06 | 0.20 | 0.56 | 72.75 | 66.39 | 75.93 | 0.30 | MUFOLD−Server | 29 | ||
| 30 | BioSerf | 72.34 | 64.14 | 75.30 | 0.02 | 0.48 | 72.34 | 64.14 | 75.30 | 0.16 | BioSerf | 30 | |||
| 31 | keasar−server | 72.13 | 64.89 | 71.44 | 0.04 | 75.20 | 67.83 | 72.37 | 0.11 | keasar−server | 31 | ||||
| 32 | PSI | 72.13 | 63.93 | 78.03 | 0.86 | 77.66 | 71.24 | 78.03 | 0.73 | 0.68 | 0.78 | PSI | 32 | ||
| 33 | pipe_int | 71.93 | 66.19 | 75.90 | 0.26 | 0.56 | 75.61 | 68.58 | 75.90 | 0.21 | 0.11 | 0.30 | pipe_int | 33 | |
| 34 | 3D−JIGSAW_V3 | 71.93 | 61.95 | 78.31 | 0.90 | 71.93 | 64.48 | 78.32 | 0.84 | 3D−JIGSAW_V3 | 34 | ||||
| 35 | GS−MetaServer2 | 71.72 | 65.03 | 73.11 | 0.06 | 0.17 | 76.03 | 69.19 | 75.47 | 0.32 | 0.24 | 0.20 | GS−MetaServer2 | 35 | |
| 36 | mGenTHREADER | 71.72 | 62.98 | 75.57 | 0.52 | 71.72 | 62.98 | 75.57 | 0.22 | mGenTHREADER | 36 | ||||
| 37 | FALCON | 71.31 | 62.43 | 73.40 | 0.22 | 71.31 | 62.43 | 73.40 | FALCON | 37 | |||||
| 38 | SAM−T06−server | 71.11 | 65.78 | 75.39 | 0.19 | 0.49 | 75.82 | 70.63 | 75.39 | 0.26 | 0.55 | 0.18 | SAM−T06−server | 38 | |
| 39 | HHpred2 | 71.11 | 63.73 | 77.92 | 0.85 | 71.11 | 63.73 | 77.92 | 0.75 | HHpred2 | 39 | ||||
| 40 | FFASflextemplate | 71.11 | 60.72 | 69.29 | 71.72 | 62.84 | 70.13 | FFASflextemplate | 40 | ||||||
| 41 | MULTICOM−RANK | 70.90 | 65.98 | 76.35 | 0.22 | 0.63 | 73.36 | 69.13 | 76.44 | 0.23 | 0.42 | MULTICOM−RANK | 41 | ||
| 42 | nFOLD3 | 70.90 | 65.85 | 71.46 | 0.20 | 71.11 | 65.85 | 71.85 | nFOLD3 | 42 | |||||
| 43 | FUGUE_KM | 70.90 | 62.30 | 70.17 | 71.31 | 62.30 | 70.17 | FUGUE_KM | 43 | ||||||
| 44 | COMA | 70.70 | 64.55 | 68.37 | 70.70 | 66.87 | 77.84 | 0.74 | COMA | 44 | |||||
| 45 | forecast | 70.70 | 63.87 | 67.99 | 76.43 | 67.83 | 73.38 | 0.42 | forecast | 45 | |||||
| 46 | BAKER−ROBETTA | 70.29 | 63.18 | 69.65 | 73.36 | 66.87 | 72.61 | BAKER−ROBETTA | 46 | ||||||
| 47 | rehtnap | 70.29 | 59.49 | 65.51 | 70.29 | 59.49 | 65.51 | rehtnap | 47 | ||||||
| 48 | Distill | 70.08 | 64.34 | 64.77 | 70.90 | 66.46 | 65.61 | Distill | 48 | ||||||
| 49 | FEIG | 69.67 | 60.52 | 72.85 | 0.14 | 78.48 | 72.88 | 77.60 | 0.93 | 1.04 | 0.68 | FEIG | 49 | ||
| 50 | FAMSD | 69.47 | 63.32 | 63.16 | 74.80 | 69.81 | 72.82 | 0.01 | 0.37 | FAMSD | 50 | ||||
| 51 | Poing | 68.85 | 60.52 | 73.43 | 0.22 | 73.77 | 66.53 | 77.87 | 0.74 | Poing | 51 | ||||
| 52 | Phyre2 | 68.85 | 60.52 | 73.43 | 0.22 | 73.77 | 66.53 | 77.87 | 0.74 | Phyre2 | 52 | ||||
| 53 | Phragment | 68.85 | 60.52 | 73.43 | 0.22 | 73.77 | 66.53 | 77.87 | 0.74 | Phragment | 53 | ||||
| 54 | SAM−T02−server | 68.65 | 60.18 | 65.16 | 73.36 | 65.57 | 71.72 | SAM−T02−server | 54 | ||||||
| 55 | Pcons_local | 68.44 | 63.66 | 68.46 | 73.57 | 67.01 | 71.23 | Pcons_local | 55 | ||||||
| 56 | HHpred4 | 68.24 | 60.59 | 74.89 | 0.42 | 68.24 | 60.59 | 74.89 | 0.07 | HHpred4 | 56 | ||||
| 57 | ACOMPMOD | 67.01 | 57.85 | 70.74 | 77.46 | 67.62 | 73.43 | 0.68 | ACOMPMOD | 57 | |||||
| 58 | mariner1 | 65.57 | 57.38 | 57.60 | 77.25 | 68.24 | 73.72 | 0.62 | 0.04 | mariner1 | 58 | ||||
| 59 | CpHModels | 65.37 | 59.63 | 65.22 | 65.37 | 59.63 | 65.22 | CpHModels | 59 | ||||||
| 60 | huber−torda−server | 63.32 | 61.13 | 55.34 | 63.32 | 61.13 | 55.34 | huber−torda−server | 60 | ||||||
| 61 | Pushchino | 54.71 | 44.47 | 45.29 | 54.71 | 44.47 | 45.29 | Pushchino | 61 | ||||||
| 62 | FOLDpro | 49.80 | 41.73 | 43.44 | 63.93 | 56.83 | 62.55 | FOLDpro | 62 | ||||||
| 63 | Fiser−M4T | 38.32 | 37.09 | 21.71 | 38.32 | 37.09 | 21.71 | Fiser−M4T | 63 | ||||||
| 64 | RBO−Proteus | 34.84 | 29.64 | 41.88 | 34.84 | 29.64 | 43.93 | RBO−Proteus | 64 | ||||||
| 65 | MUFOLD−MD | 29.10 | 22.95 | 41.02 | 30.94 | 26.43 | 46.87 | MUFOLD−MD | 65 | ||||||
| 66 | FALCON_CONSENSUS | 26.23 | 22.54 | 38.93 | 71.31 | 62.43 | 73.40 | FALCON_CONSENSUS | 66 | ||||||
| 67 | OLGAFS | 17.42 | 14.69 | 12.19 | 27.46 | 24.59 | 20.31 | OLGAFS | 67 | ||||||
| 68 | schenk−torda−server | 15.98 | 14.21 | 18.96 | 17.21 | 14.41 | 21.26 | schenk−torda−server | 68 | ||||||
| 69 | RANDOM | 15.83 | 13.24 | 15.83 | 15.83 | 13.24 | 15.83 | RANDOM | 69 | ||||||
| 70 | mahmood−torda−server | 13.93 | 10.66 | 17.94 | 17.01 | 14.34 | 23.52 | mahmood−torda−server | 70 | ||||||
| 71 | 3Dpro | 3Dpro | 71 | ||||||||||||
| 72 | BHAGEERATH | BHAGEERATH | 72 | ||||||||||||
| 73 | YASARA | YASARA | 73 | ||||||||||||
| 74 | test_http_server_01 | test_http_server_01 | 74 | ||||||||||||