T0510
Fibrinogen binding protein from Staphylococcus aureus
Target sequence:
>T0510 Fibrinogen binding protein, Staphylococcus aureus, 288 residues
MAYDGLFTKKMVESLQFLTTGRVHKINQPDNDTILMVVRQNRQNHQLLLSIHPNFSRLQLTTKKYDNPFNPPMFARVFRKHLEGGIIESIKQIGNDRRIEIDIKSKDEIGDTIYRTVILEIMGKHSNLILVDENRKIIEGFKHLTPNTNHYRTVMPGFNYEAPPTQHKINPYDITGAEVLKYIDFNAGNIAKQLLNQFEGFSPLITNEIVSRRQFMTSSTLPEAFDEVMAETKLPPTPIFHKNHETGKEDFYFIKLNQFNDDTVTYDSLNDLLDRFYDARGERERVKQ
Structure:
Determined by:
MCSG
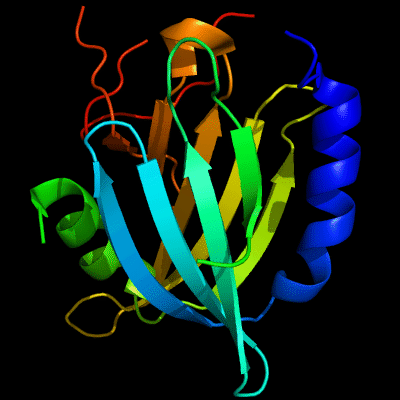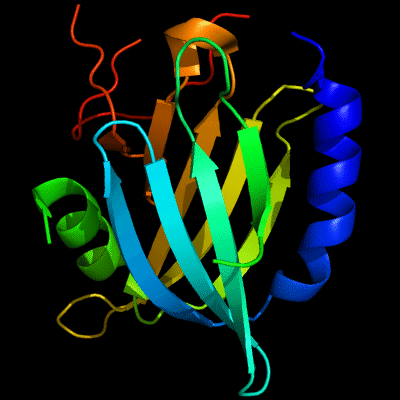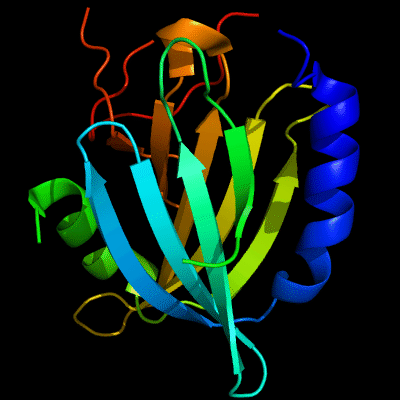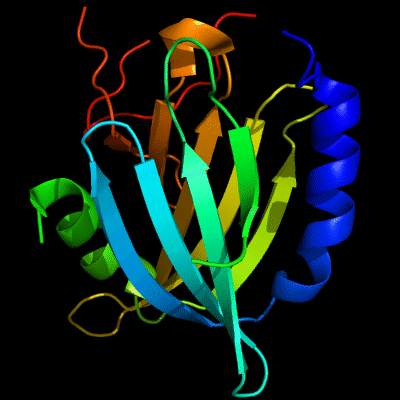
PDB ID: 3doa
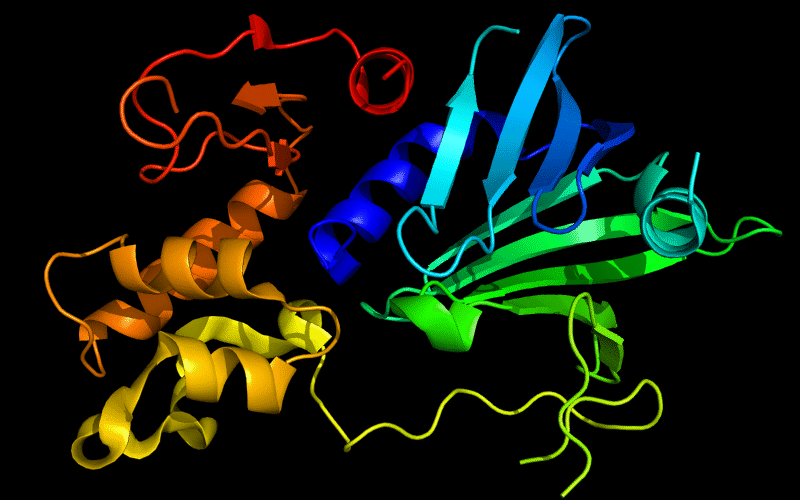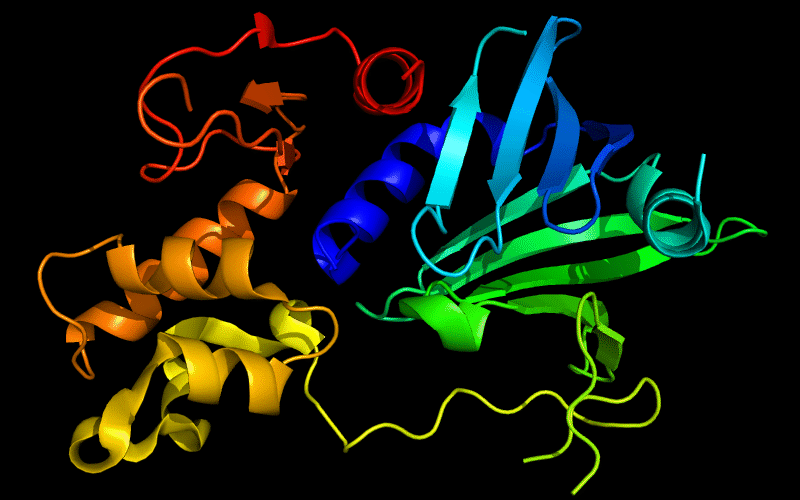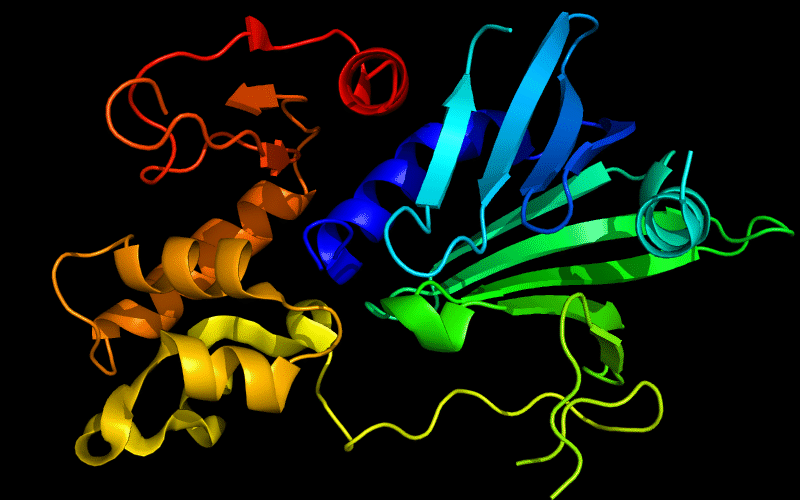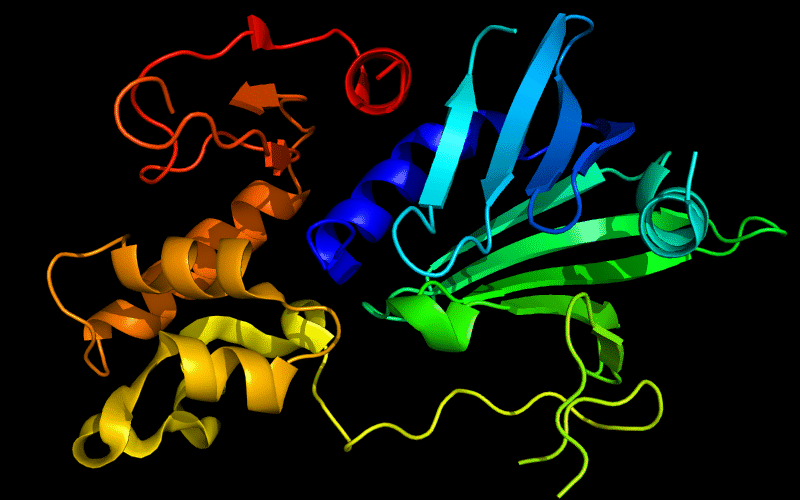
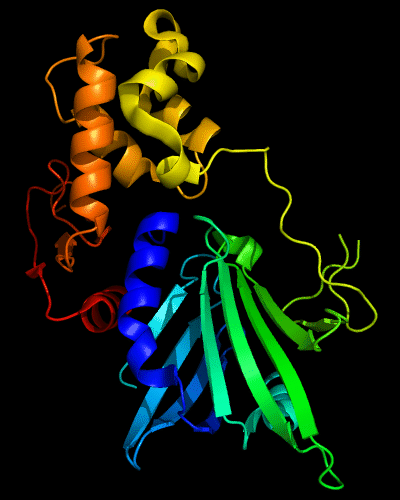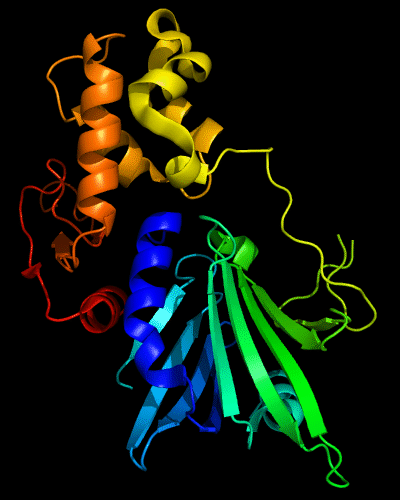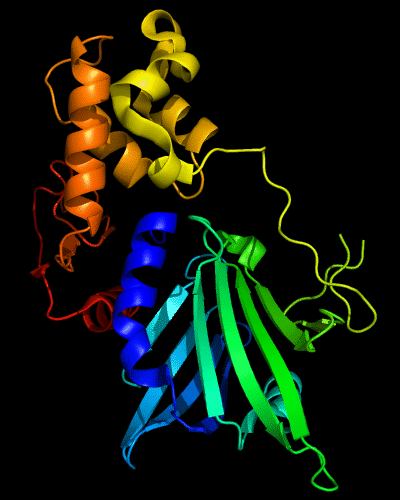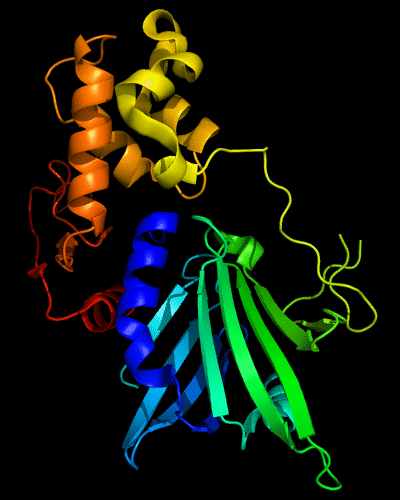
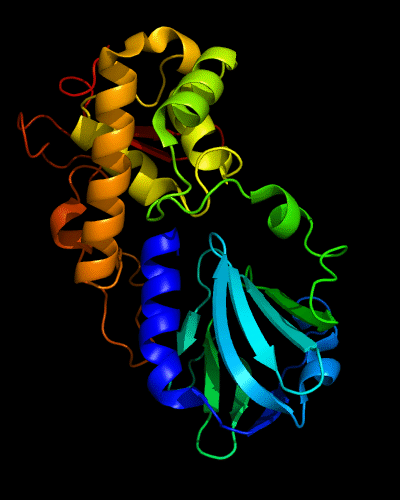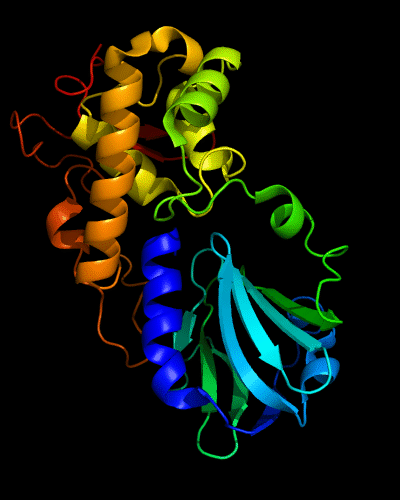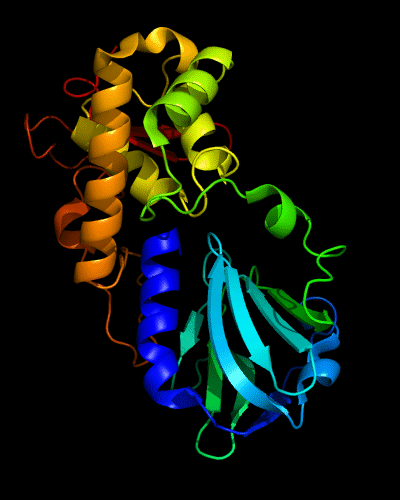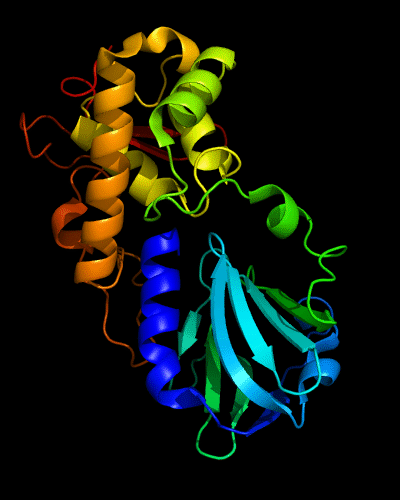
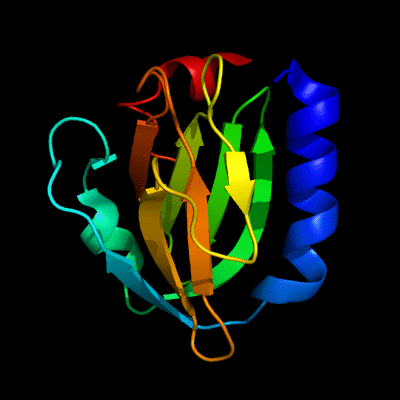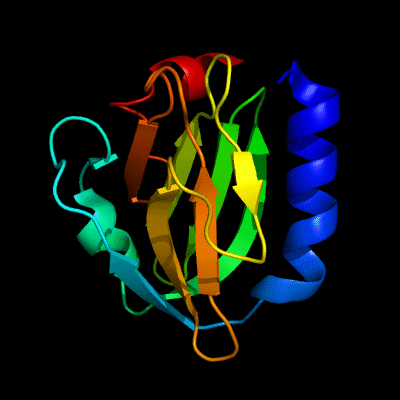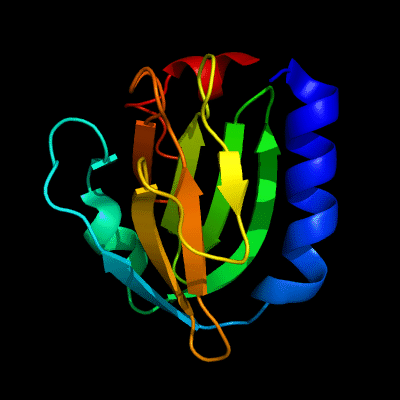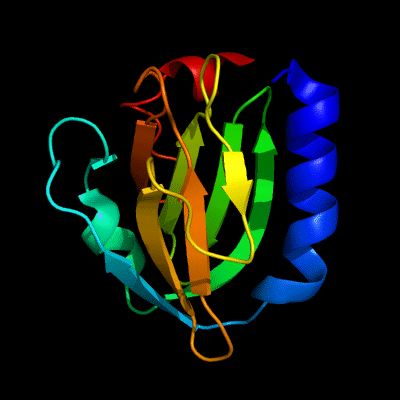
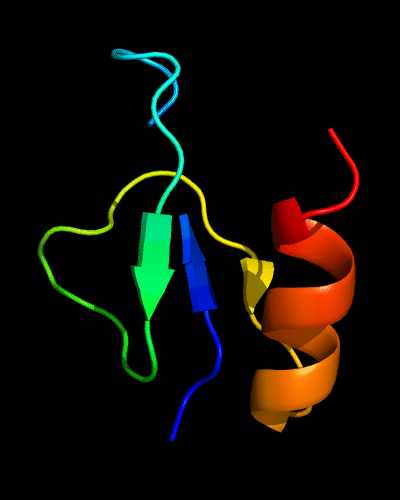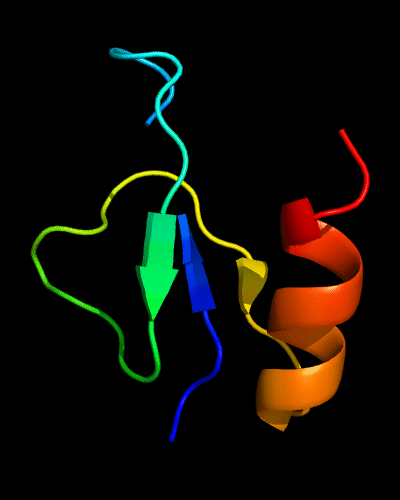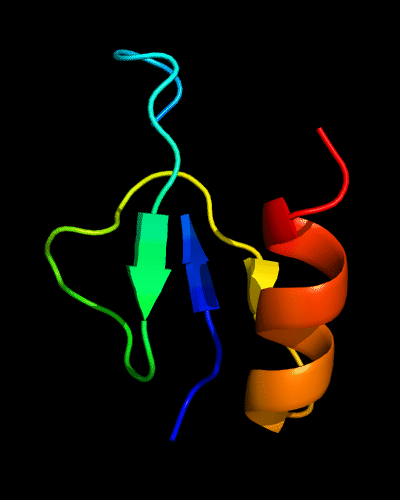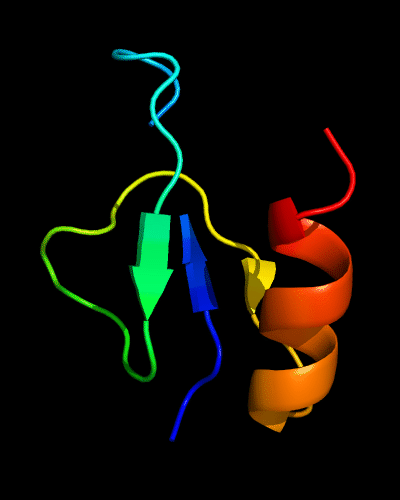
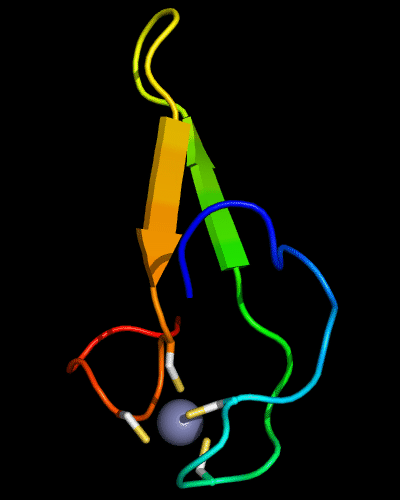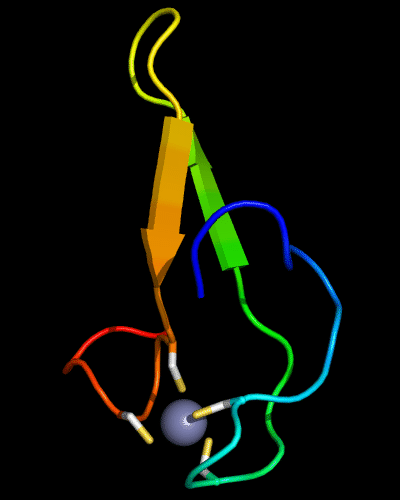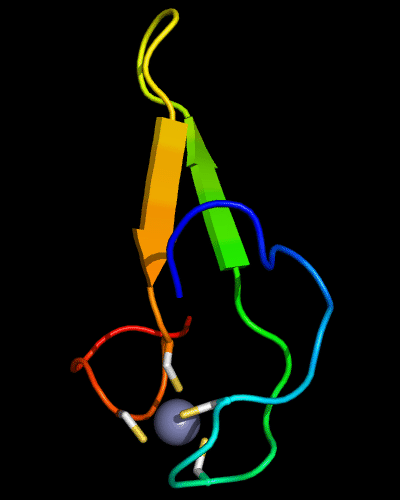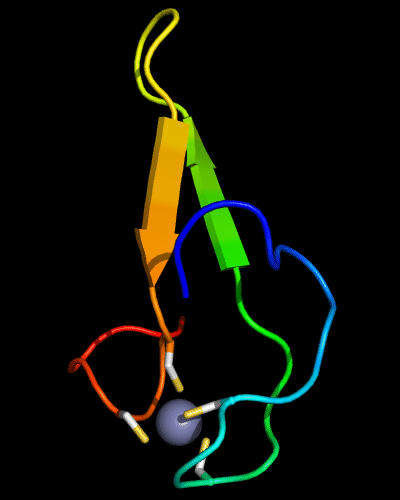
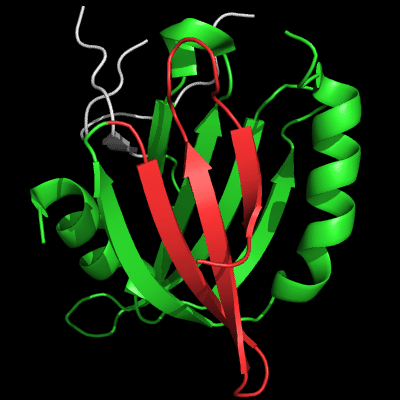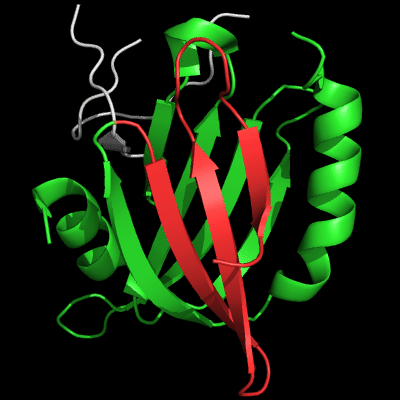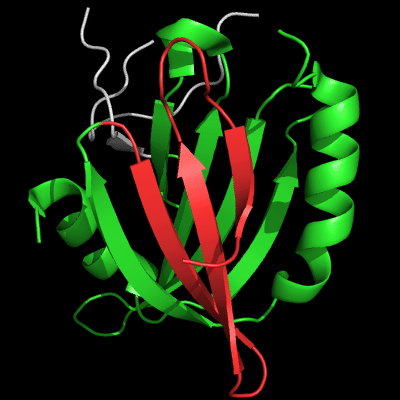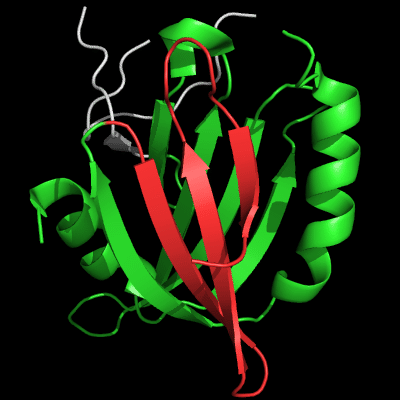
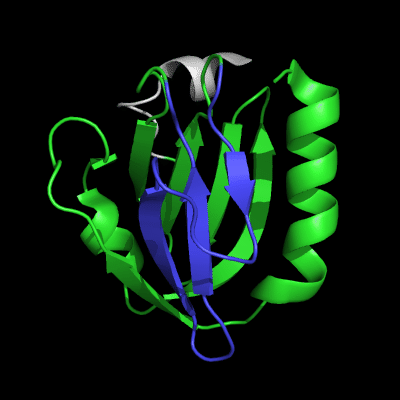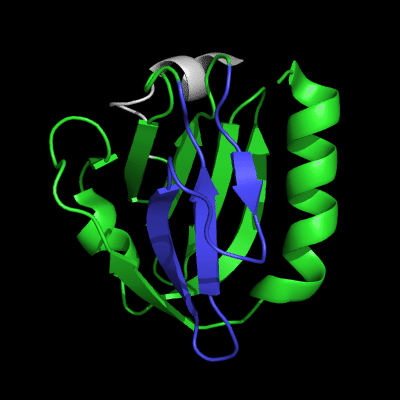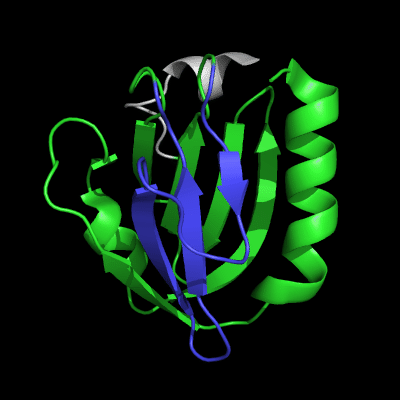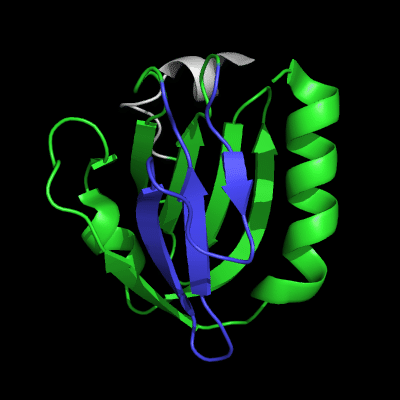
Cartoon diagram of 510: 3doa
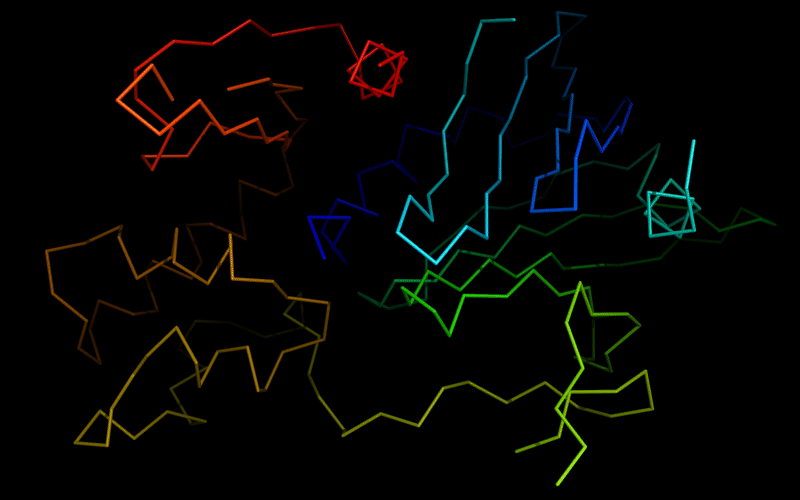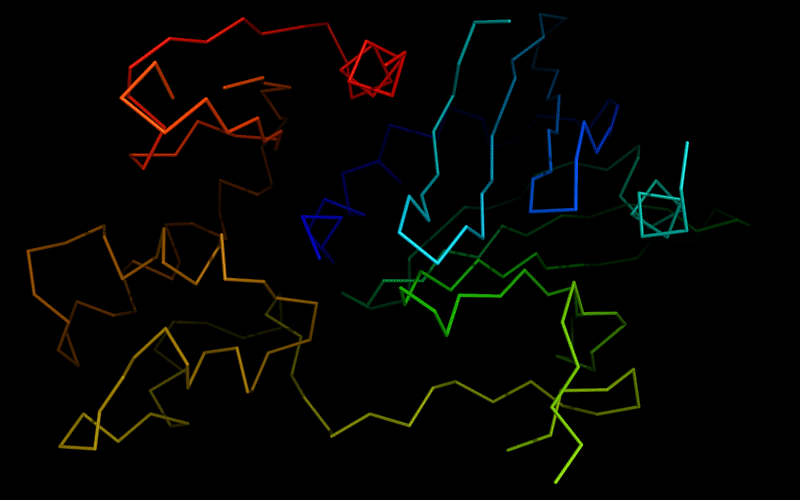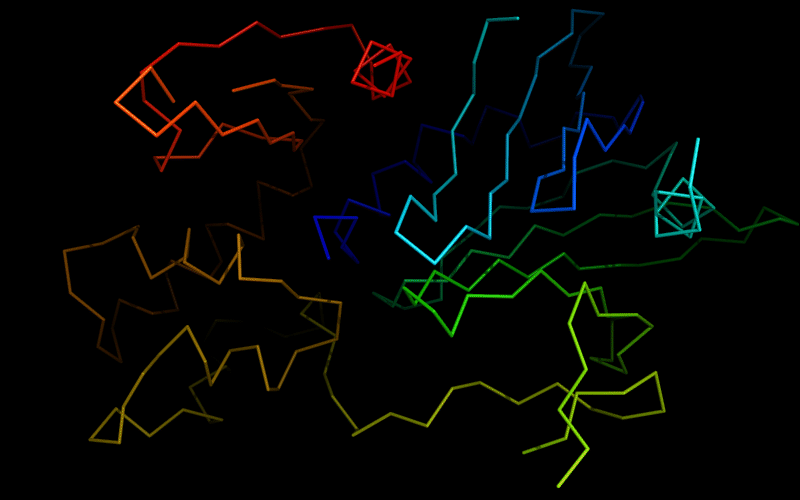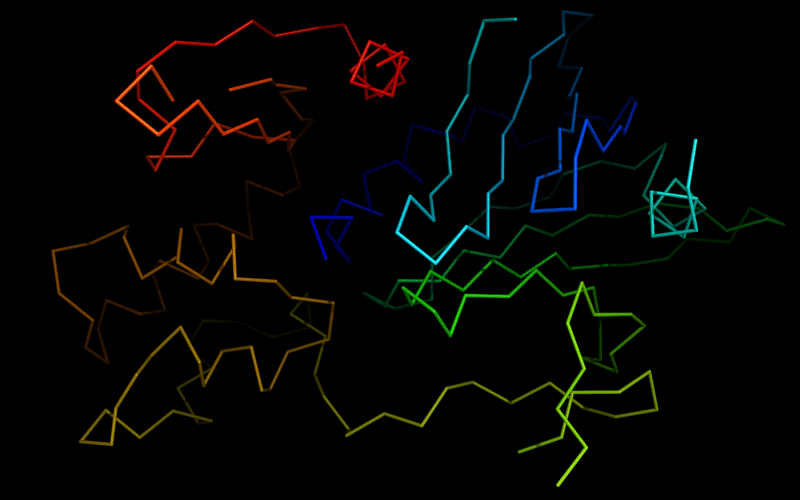
Ribbon diagram of 510: 3doa
Domains: PyMOL of domains
Three domains. Residue ranges in PDB: 1-165; 166-235 and 236-279. Residue ranges in target:1-165; 166-235 and 236-279.
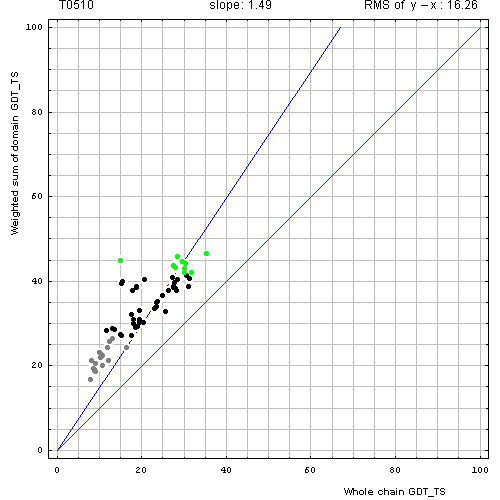
Correlation between weighted by the number of residues sum of GDT-TS scores for domain-based evlatuation (y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
To compute the weighted sum, GDT-TS for each domain was multiplied by the domain length, and this sum was divided by the sum of domain lengths. Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the weighted sum of domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
N-domain: N-terminal domain of MutM-like DNA repair proteins fold.
Middle domain: S13-like H2TH domain fold.
C-domain: deteriorated treble-clef finger with Glucocorticoid receptor-like (DNA-binding domain) fold.
N and C-domains experience amazing structural transformations compared to homologous protein MutM. Apparently, T0510 and MutM are homologous throughout the chain in all three domains, and even relative positions of these domains appears very similar:
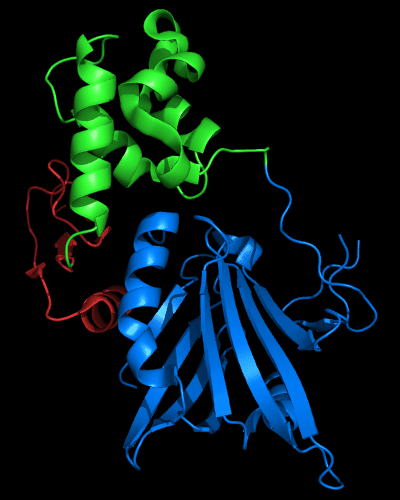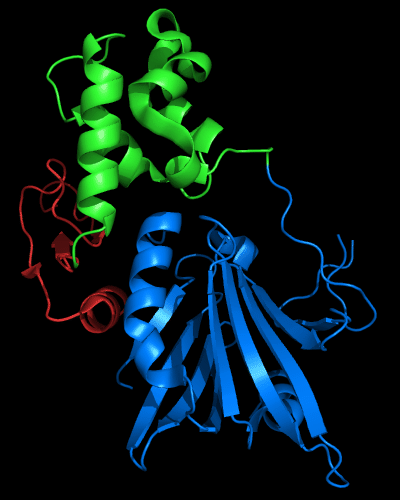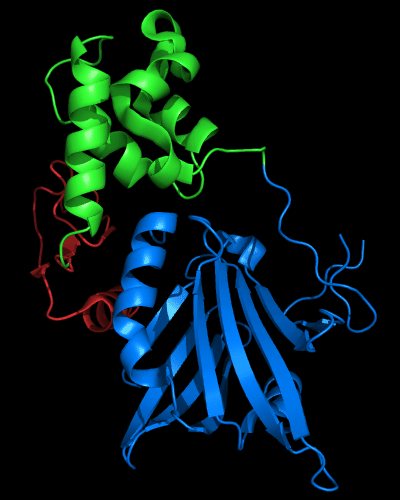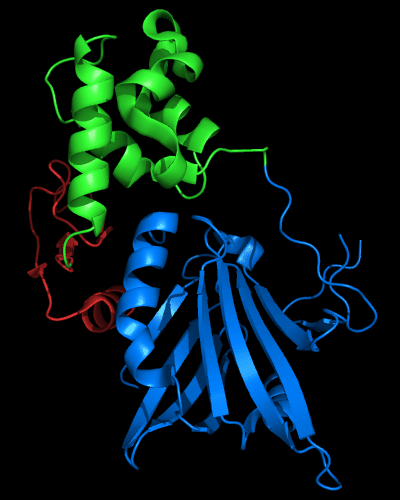
Cartoon diagram of 510 domains: 3doa, N-, middle and C-domains
are shown in
blue, green and red, respectively
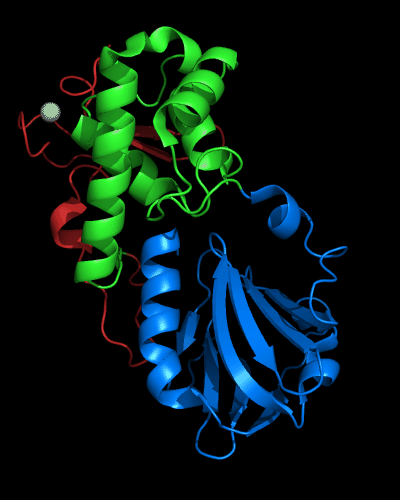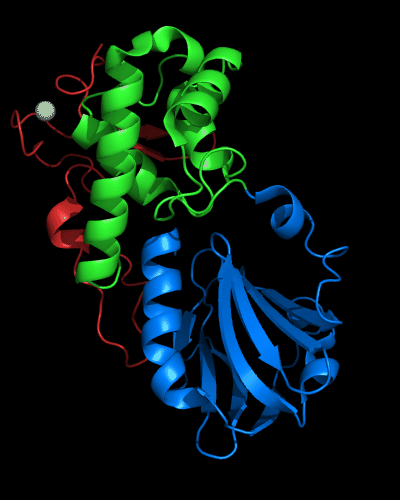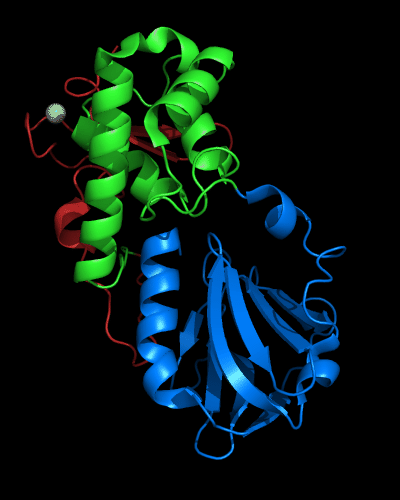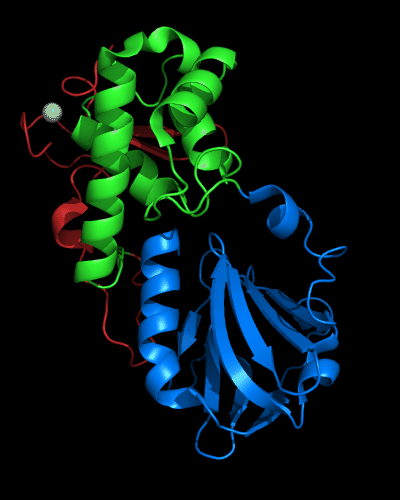
Cartoon diagram of MutM domains: 1ee8_A, N-, middle and C-domains
are shown in
blue, green and red, respectively,
Zn ion is shown as a white ball.
However, closer look at the N-domains reveals large topological differences. The view of N-domain is "from the back" compared to the images above:
Architecture appears similar between the two proteins: two β-sheets flanked by 2 α-helices on the sides, but topology is quite different. The β-sheet facing the viewer is N-terminal (blue-cyan-green) in T0510, and is inserted in the middle of the molecule (yellow-orange) in MutM. Common superimposable parts of both molecules are shown in green below, different insertions are colored red and blue.
N-domain of 510: 3doa residues 1-165
insertion close to the N-terminus is red
N-domain of MutM: 1ee8 chain A residues 1-121
insertion in the middle of the domain is blue
T0510 has a 3-stranded insertion (red) after the N-terminal α-helix, and MutM has a 3-stranded insertion (blue) after the 4th common β-strand. Although both insertions form 3-stranded antiparallel β-sheets, they are different in topology: meander in T0510, and βLββ in MutM. Insertions are not surprising in remote homologs. Convergence to similar architecture through independent insertions/deletions is not very common.
Despite this interesting peculiarity, there is no doubt about homology between N-domains of these proteins. COMPASS being queried with just the N-terminal domain sequence, without a more similar H2TH middle domain, finds the first MutM protein as 16th hit. Here is an alignment with one of MutM proteins found by COMPASS. This alignment is correct in the middle region.
Subject = d1ee8a2 b.113.1.1 DNA repair protein MutM (Fpg) length=121 filtered_length=121 Neff=15.190 Smith-Waterman score = 47 Evalue = 1.15e+01 QUERY 59 QLTTKKYDNPFNPPMFARVFRKHLEGGIIESIK========QIGNDRRIEIDIKSKDEIGDTIYRTVILEIMGKHSNLILVD 132 CONSENSUS_1 59 HLTEFPRENPKTPSNFAMLLRKHLKGARLVSIE========QLGFDRILELEFGSGDELGDEILYRLILELFGRHGNIILLD 132 +L+ ++ ++ + +++ + ++L+G+R++S+E L+ + +L +++G L+ + +++ ++L+ +G ++++D CONSENSUS_2 16 LLVGRTITRVEVRWPKPADFAAALVGRRILSVERRGKYLLLELDGGLVLVVHLGMSGRLRLVPPKHDHVRLELDGGELRFVD 97 d1ee8a2 16 LVLGQTLRQVVHRDPARYRNTALAEGRRILEVDRRGKFLLFALEGGVELVAHLGMTGGFRLEPTPHTRAALVLEGRTLYFHD 97
The C-domain in T0510 is a treble-clef finger that lost its Zn-binding site, deteriorated beyond recognition and gained the C-terminal β/α-unit:
The blue and cyan β-strands in T0510 are structurally equivalent and homologous to green and orange β-strands in MutM, respectively.
CASP category:
Whole chain: Fold recognition.
1st domain: Fold recognition. 2nd domain: Comparative modeling:hard. 3rd domain: Fold recognition.
Closest templates:
Target sequence - PDB file inconsistencies:
T0510 3doa.pdb T0510.pdb PyMOL PyMOL of domains
T0510 1 MAYDGLFTKKMVESLQFLTTGRVHKINQPDNDTILMVVRQNRQNHQLLLSIHPNFSRLQLTTKKYDNPFNPPMFARVFRKHLEGGIIESIKQIGNDRRIEIDIKSKDEIGDTIYRTVILEIMGKHSNLILVDENRKIIEGFKHLTPNTNHYRTVMPGFNYEAPPTQHKINPYDITGAEVLKYIDFNAGNIAKQLLNQFEGFSPLITNEIVSRRQFMTSSTLPEAFDEVMAETKLPPTPIFHKNHETGKEDFYFIKLNQFNDDTVTYDSLNDLLDRFYDARGERERVKQ 288 |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~|||||||||||||||||||||||||||||||||||||~~~~~~~~~ 3doaA 1 MAYDGLFTKKMVESLQFLTTGRVHKINQPDNDTILMVVRQNRQNHQLLLSIHPNFSRLQLTTK-------PPMFARVFRKHLEGGIIESIKQIGNDRRIEIDIKSKDEIGDTIYRTVILEIMGKHSNLILVDENRKIIEGFKHLTP-----RTVMPGFNYEAPPTQHKINPYDITGAEVLKYIDFNAGNIAKQLLNQFEGFSPLITNEIVSRRQFMTSSTLPEAFDEVMAETKLPPTPIFH-NHETGKEDFYFIKLNQFNDDTVTYDSLNDLLDRFYDA--------- 279
3-domain protein, all domains used in evaluation:
1st domain: target 1-63, 71-146, 152-165 ; pdb 1-63, 71-146, 152-165
2nd domain: target 166-235 ; pdb 166-235
3rd domain: target 236-241, 243-279 ; pdb 236-241, 243-279
Sequence classification:
Fibronectin-binding protein A N-terminus (FbpA) in Pfam.
Server predictions:
T0510:pdb 1-279:seq 1-279:FR; alignment
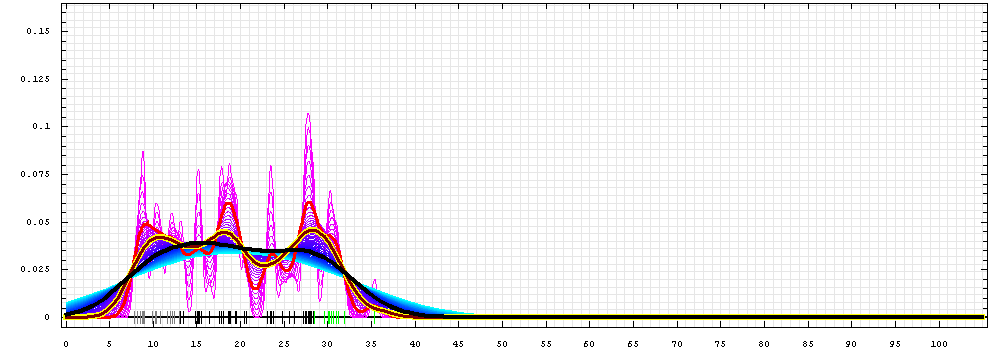
First models for T0510:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
510_1:pdb 1-165:seq 1-165:FR; alignment
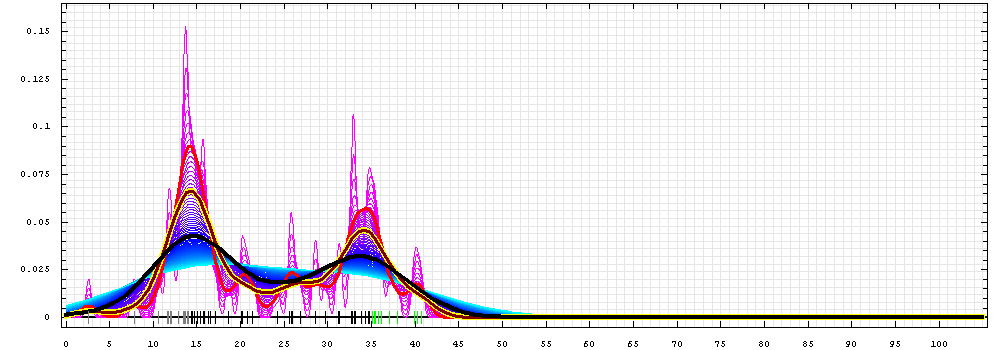
First models for T0510_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
510_2:pdb 166-235:seq 166-235:CM_hard; alignment
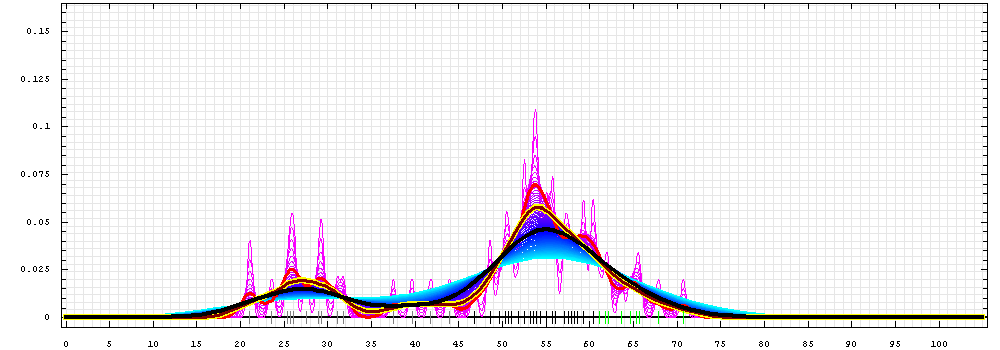
First models for T0510_2:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
510_3:pdb 236-279:seq 236-279:FR; alignment
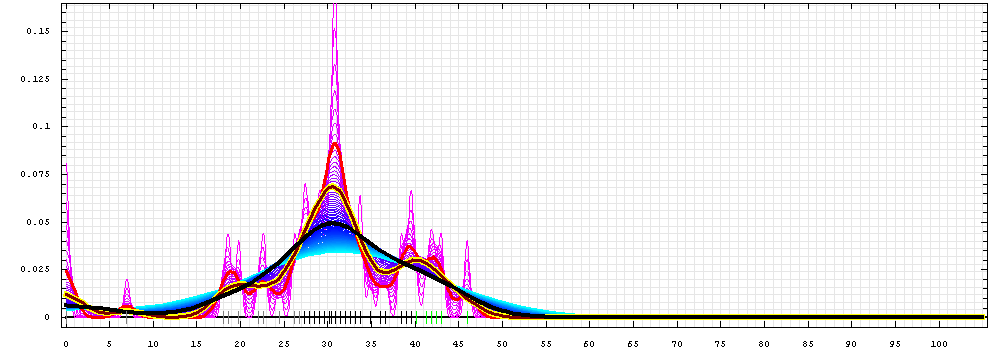
First models for T0510_3:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0510 | 510_1 | 510_2 | 510_3 | T0510 | 510_1 | 510_2 | 510_3 | T0510 | 510_1 | 510_2 | 510_3 | T0510 | 510_1 | 510_2 | 510_3 | ||||||||||||||||||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | SAM−T08−server | 35.34 | 28.51 | 36.99 | 40.20 | 36.38 | 39.32 | 65.36 | 53.93 | 56.88 | 38.37 | 37.02 | 41.56 | 2.14 | 2.16 | 1.60 | 1.71 | 1.83 | 1.67 | 1.36 | 0.87 | 1.04 | 0.99 | 1.43 | 0.66 | 35.62 | 28.51 | 36.99 | 41.18 | 37.91 | 40.39 | 67.50 | 53.93 | 59.77 | 38.37 | 37.02 | 41.56 | 1.86 | 1.80 | 1.44 | 1.74 | 1.89 | 1.54 | 1.28 | 0.43 | 0.96 | 0.52 | 0.90 | 0.18 | SAM−T08−server | 1 |
| 2 | HHpred2 | 31.86 | 23.65 | 35.77 | 35.46 | 31.48 | 36.14 | 65.71 | 53.57 | 63.35 | 27.33 | 25.97 | 32.28 | 1.62 | 1.21 | 1.44 | 1.18 | 1.23 | 1.28 | 1.40 | 0.83 | 1.92 | 31.86 | 23.65 | 35.77 | 35.46 | 31.48 | 36.14 | 65.71 | 53.57 | 63.35 | 27.33 | 25.97 | 32.28 | 1.26 | 0.80 | 1.27 | 0.99 | 0.99 | 0.96 | 1.05 | 0.38 | 1.49 | HHpred2 | 2 | ||||||
| 3 | HHpred5 | 31.20 | 23.97 | 36.30 | 31.37 | 25.16 | 35.61 | 67.86 | 55.71 | 65.44 | 29.07 | 27.13 | 34.95 | 1.52 | 1.28 | 1.51 | 0.74 | 0.46 | 1.22 | 1.63 | 1.09 | 2.20 | 31.20 | 23.97 | 36.30 | 31.37 | 25.16 | 35.61 | 67.86 | 55.71 | 65.44 | 29.07 | 27.13 | 34.95 | 1.15 | 0.87 | 1.34 | 0.46 | 0.12 | 0.88 | 1.33 | 0.71 | 1.80 | HHpred5 | 3 | ||||||
| 4 | HHpred4 | 31.02 | 25.47 | 31.01 | 26.80 | 21.46 | 26.66 | 70.71 | 58.33 | 66.38 | 28.49 | 24.61 | 33.68 | 1.49 | 1.57 | 0.84 | 0.23 | 0.01 | 0.11 | 1.93 | 1.40 | 2.33 | 31.02 | 25.47 | 31.01 | 26.80 | 21.46 | 26.66 | 70.71 | 58.33 | 66.38 | 28.49 | 24.61 | 33.68 | 1.12 | 1.18 | 0.58 | 1.71 | 1.11 | 1.93 | HHpred4 | 4 | |||||||||
| 5 | COMA−M | 30.64 | 25.85 | 32.93 | 36.11 | 34.15 | 34.93 | 59.29 | 52.62 | 53.35 | 30.81 | 25.39 | 35.04 | 1.44 | 1.64 | 1.08 | 1.26 | 1.56 | 1.13 | 0.71 | 0.72 | 0.56 | 30.64 | 25.85 | 32.93 | 36.11 | 34.15 | 34.93 | 59.64 | 55.59 | 55.82 | 31.98 | 28.30 | 37.28 | 1.06 | 1.25 | 0.85 | 1.08 | 1.37 | 0.79 | 0.24 | 0.69 | 0.39 | COMA−M | 5 | ||||||
| 6 | pro−sp3−TASSER | 30.36 | 25.38 | 40.07 | 37.91 | 34.53 | 40.61 | 60.36 | 57.50 | 58.70 | 39.53 | 34.50 | 50.46 | 1.39 | 1.55 | 1.99 | 1.45 | 1.60 | 1.83 | 0.82 | 1.30 | 1.29 | 1.17 | 1.03 | 1.63 | 30.36 | 25.38 | 40.07 | 37.91 | 34.53 | 42.58 | 60.36 | 57.50 | 58.70 | 45.35 | 41.09 | 59.15 | 1.02 | 1.16 | 1.89 | 1.31 | 1.42 | 1.84 | 0.34 | 0.98 | 0.81 | 1.77 | 1.57 | 2.41 | pro−sp3−TASSER | 6 |
| 7 | Zhang−Server | 30.17 | 22.15 | 38.80 | 34.64 | 30.83 | 39.51 | 63.57 | 60.00 | 63.13 | 33.72 | 24.03 | 41.14 | 1.37 | 0.92 | 1.83 | 1.09 | 1.15 | 1.70 | 1.17 | 1.60 | 1.89 | 0.26 | 0.62 | 34.68 | 27.00 | 45.39 | 42.48 | 36.66 | 46.99 | 67.14 | 60.00 | 67.90 | 35.47 | 31.20 | 44.33 | 1.71 | 1.49 | 2.66 | 1.90 | 1.71 | 2.45 | 1.23 | 1.37 | 2.16 | 0.01 | 0.53 | Zhang−Server | 7 | ||
| 8 | CpHModels | 30.07 | 22.84 | 29.44 | 35.29 | 29.19 | 33.56 | 64.64 | 52.98 | 58.85 | 19.77 | 16.67 | 13.66 | 1.35 | 1.06 | 0.64 | 1.17 | 0.95 | 0.96 | 1.28 | 0.76 | 1.31 | 30.07 | 22.84 | 29.44 | 35.29 | 29.19 | 33.56 | 64.64 | 52.98 | 58.85 | 19.77 | 16.67 | 13.66 | 0.97 | 0.64 | 0.35 | 0.97 | 0.68 | 0.60 | 0.90 | 0.29 | 0.83 | CpHModels | 8 | ||||||
| 9 | RAPTOR | 29.61 | 23.97 | 35.21 | 39.87 | 36.49 | 37.32 | 53.93 | 47.02 | 47.36 | 45.93 | 38.57 | 56.60 | 1.28 | 1.28 | 1.37 | 1.67 | 1.84 | 1.43 | 0.13 | 0.04 | 2.18 | 1.68 | 2.30 | 29.89 | 24.69 | 35.64 | 39.87 | 36.49 | 37.32 | 53.93 | 47.02 | 48.14 | 45.93 | 44.38 | 59.86 | 0.94 | 1.02 | 1.25 | 1.57 | 1.69 | 1.12 | 1.87 | 2.12 | 2.50 | RAPTOR | 9 | ||||
| 10 | COMA | 28.48 | 23.93 | 31.70 | 33.17 | 30.12 | 31.67 | 61.79 | 50.36 | 58.22 | 30.81 | 28.30 | 35.42 | 1.11 | 1.27 | 0.93 | 0.93 | 1.07 | 0.73 | 0.97 | 0.44 | 1.22 | 0.05 | 29.61 | 26.10 | 32.93 | 34.97 | 30.12 | 34.95 | 61.79 | 56.07 | 58.22 | 30.81 | 28.30 | 37.28 | 0.90 | 1.30 | 0.85 | 0.93 | 0.81 | 0.79 | 0.53 | 0.76 | 0.74 | COMA | 10 | |||||
| 11 | fais−server | 28.38 | 23.87 | 34.66 | 40.69 | 34.59 | 34.36 | 59.29 | 48.09 | 59.12 | 41.86 | 40.12 | 47.78 | 1.10 | 1.26 | 1.30 | 1.76 | 1.61 | 1.06 | 0.71 | 0.17 | 1.34 | 1.54 | 1.93 | 1.34 | 30.83 | 25.50 | 35.51 | 40.69 | 34.59 | 36.37 | 67.14 | 54.76 | 59.12 | 41.86 | 40.12 | 50.18 | 1.09 | 1.18 | 1.23 | 1.67 | 1.43 | 0.99 | 1.23 | 0.56 | 0.87 | 1.15 | 1.41 | 1.27 | fais−server | 11 |
| 12 | BAKER−ROBETTA | 28.10 | 22.65 | 26.34 | 33.01 | 29.96 | 28.18 | 53.57 | 47.38 | 46.29 | 29.07 | 26.36 | 29.55 | 1.05 | 1.02 | 0.24 | 0.92 | 1.05 | 0.30 | 0.09 | 0.09 | 28.10 | 22.65 | 27.76 | 33.01 | 29.96 | 29.94 | 69.29 | 59.52 | 62.80 | 39.53 | 33.33 | 46.11 | 0.66 | 0.60 | 0.10 | 0.68 | 0.78 | 0.10 | 1.52 | 1.30 | 1.41 | 0.73 | 0.29 | 0.75 | BAKER−ROBETTA | 12 | ||||
| 13 | Phyre_de_novo | 28.01 | 20.55 | 31.71 | 37.09 | 30.34 | 34.78 | 60.36 | 53.45 | 50.44 | 36.63 | 33.14 | 42.66 | 1.04 | 0.61 | 0.93 | 1.36 | 1.09 | 1.11 | 0.82 | 0.82 | 0.17 | 0.72 | 0.82 | 0.78 | 29.04 | 21.08 | 33.39 | 38.40 | 31.75 | 36.49 | 63.57 | 54.52 | 55.05 | 44.19 | 41.47 | 46.26 | 0.81 | 0.28 | 0.92 | 1.38 | 1.03 | 1.01 | 0.76 | 0.52 | 0.27 | 1.56 | 1.64 | 0.77 | Phyre_de_novo | 13 |
| 14 | GS−KudlatyPred | 27.91 | 22.71 | 26.67 | 33.01 | 28.87 | 28.77 | 53.93 | 47.26 | 48.17 | 30.81 | 28.88 | 30.45 | 1.03 | 1.03 | 0.29 | 0.92 | 0.91 | 0.37 | 0.13 | 0.07 | 0.14 | 27.91 | 22.71 | 26.83 | 33.50 | 31.86 | 29.02 | 55.71 | 48.81 | 48.76 | 31.98 | 29.26 | 30.80 | 0.63 | 0.61 | 0.74 | 1.05 | GS−KudlatyPred | 14 | |||||||||||
| 15 | 3D−JIGSAW_V3 | 27.73 | 22.52 | 21.56 | 33.82 | 28.92 | 25.61 | 57.14 | 52.62 | 51.04 | 18.61 | 15.50 | 14.68 | 1.00 | 1.00 | 1.00 | 0.92 | 0.48 | 0.72 | 0.25 | 27.73 | 22.52 | 21.58 | 33.82 | 30.28 | 25.98 | 57.14 | 52.62 | 51.10 | 18.61 | 17.83 | 15.11 | 0.60 | 0.57 | 0.78 | 0.83 | 0.23 | 3D−JIGSAW_V3 | 15 | ||||||||||||
| 16 | nFOLD3 | 27.54 | 20.90 | 19.90 | 34.80 | 32.84 | 24.68 | 57.50 | 52.50 | 53.60 | 0.00 | 0.00 | 5.09 | 0.97 | 0.68 | 1.11 | 1.40 | 0.51 | 0.70 | 0.60 | 28.38 | 23.25 | 19.94 | 35.95 | 34.75 | 24.68 | 58.93 | 55.95 | 53.60 | 40.12 | 34.30 | 44.47 | 0.70 | 0.72 | 1.06 | 1.45 | 0.15 | 0.74 | 0.06 | 0.84 | 0.45 | 0.54 | nFOLD3 | 16 | |||||||
| 17 | FEIG | 27.44 | 23.75 | 38.31 | 34.31 | 31.81 | 45.81 | 54.29 | 50.24 | 48.95 | 27.91 | 25.39 | 35.24 | 0.95 | 1.23 | 1.77 | 1.06 | 1.27 | 2.47 | 0.17 | 0.43 | 27.82 | 25.31 | 39.21 | 35.29 | 33.33 | 46.58 | 63.57 | 54.76 | 53.81 | 31.39 | 29.07 | 37.76 | 0.61 | 1.14 | 1.76 | 0.97 | 1.25 | 2.39 | 0.76 | 0.56 | 0.09 | FEIG | 17 | |||||||
| 18 | METATASSER | 27.44 | 22.24 | 31.48 | 34.64 | 26.69 | 32.81 | 50.36 | 45.36 | 49.56 | 34.88 | 31.39 | 40.50 | 0.95 | 0.94 | 0.90 | 1.09 | 0.65 | 0.87 | 0.05 | 0.44 | 0.54 | 0.55 | 29.70 | 25.25 | 42.38 | 35.13 | 31.75 | 44.76 | 58.21 | 55.83 | 60.44 | 39.53 | 35.27 | 49.79 | 0.91 | 1.13 | 2.22 | 0.95 | 1.03 | 2.14 | 0.05 | 0.73 | 1.06 | 0.73 | 0.61 | 1.22 | METATASSER | 18 | ||
| 19 | MUSTER | 27.16 | 20.46 | 31.27 | 35.13 | 29.57 | 33.57 | 59.29 | 55.83 | 53.25 | 31.98 | 26.94 | 33.43 | 0.91 | 0.60 | 0.87 | 1.15 | 1.00 | 0.96 | 0.71 | 1.10 | 0.55 | 34.96 | 26.79 | 33.57 | 35.62 | 30.23 | 33.57 | 65.71 | 55.83 | 61.35 | 31.98 | 26.94 | 33.43 | 1.75 | 1.45 | 0.95 | 1.01 | 0.82 | 0.60 | 1.05 | 0.73 | 1.20 | MUSTER | 19 | ||||||
| 20 | mGenTHREADER | 26.22 | 22.27 | 15.45 | 32.84 | 29.47 | 21.03 | 49.64 | 44.64 | 29.43 | 30.23 | 24.81 | 23.04 | 0.77 | 0.95 | 0.90 | 0.99 | 26.22 | 22.27 | 15.45 | 32.84 | 29.47 | 21.03 | 49.64 | 44.64 | 29.43 | 30.23 | 24.81 | 23.04 | 0.36 | 0.52 | 0.65 | 0.72 | mGenTHREADER | 20 | ||||||||||||||||
| 21 | 3DShot2 | 25.56 | 20.93 | 23.23 | 24.18 | 18.95 | 22.36 | 55.00 | 46.91 | 45.87 | 27.33 | 20.74 | 36.70 | 0.67 | 0.69 | 0.25 | 0.03 | 0.13 | 25.56 | 20.93 | 23.23 | 24.18 | 18.95 | 22.36 | 55.00 | 46.91 | 45.87 | 27.33 | 20.74 | 36.70 | 0.25 | 0.25 | 3DShot2 | 21 | |||||||||||||||||
| 22 | FFASstandard | 24.81 | 19.93 | 23.07 | 29.74 | 25.27 | 27.11 | 52.50 | 45.59 | 46.74 | 32.56 | 27.13 | 25.00 | 0.56 | 0.49 | 0.56 | 0.48 | 0.17 | 0.08 | 24.81 | 19.93 | 24.90 | 29.74 | 25.27 | 29.71 | 55.71 | 50.48 | 49.27 | 32.56 | 28.88 | 34.72 | 0.13 | 0.04 | 0.25 | 0.13 | 0.07 | FFASstandard | 22 | |||||||||||||
| 23 | MULTICOM−CLUSTER | 23.78 | 20.61 | 26.65 | 25.82 | 23.86 | 25.26 | 58.57 | 54.17 | 52.22 | 30.81 | 23.26 | 37.85 | 0.40 | 0.62 | 0.28 | 0.13 | 0.31 | 0.63 | 0.90 | 0.41 | 0.26 | 23.78 | 20.61 | 30.45 | 32.84 | 29.68 | 33.78 | 60.00 | 54.52 | 52.56 | 32.56 | 28.88 | 39.02 | 0.18 | 0.49 | 0.65 | 0.74 | 0.63 | 0.29 | 0.52 | MULTICOM−CLUSTER | 23 | ||||||||
| 24 | MULTICOM−REFINE | 23.59 | 20.33 | 26.69 | 25.65 | 23.91 | 25.33 | 58.21 | 53.81 | 52.20 | 30.81 | 23.64 | 37.76 | 0.37 | 0.57 | 0.29 | 0.11 | 0.31 | 0.59 | 0.86 | 0.41 | 0.25 | 32.71 | 26.19 | 31.65 | 32.68 | 29.74 | 33.63 | 61.79 | 53.81 | 57.00 | 31.98 | 26.55 | 38.98 | 1.39 | 1.32 | 0.67 | 0.63 | 0.75 | 0.61 | 0.53 | 0.41 | 0.56 | MULTICOM−REFINE | 24 | ||||||
| 25 | Pcons_dot_net | 23.40 | 19.83 | 24.32 | 28.59 | 24.56 | 24.25 | 52.50 | 48.45 | 48.15 | 22.67 | 21.90 | 29.03 | 0.35 | 0.47 | 0.43 | 0.39 | 0.21 | 24.25 | 20.61 | 24.32 | 31.37 | 27.78 | 26.72 | 57.14 | 51.67 | 53.55 | 24.42 | 22.48 | 30.57 | 0.04 | 0.18 | 0.46 | 0.48 | 0.08 | 0.05 | Pcons_dot_net | 25 | |||||||||||||
| 26 | Pcons_multi | 23.40 | 19.83 | 24.32 | 28.59 | 24.56 | 24.25 | 52.50 | 48.45 | 48.15 | 22.67 | 21.90 | 29.03 | 0.35 | 0.47 | 0.43 | 0.39 | 0.21 | 25.38 | 21.37 | 25.29 | 29.25 | 25.55 | 25.46 | 58.93 | 54.05 | 52.28 | 31.39 | 23.26 | 32.23 | 0.22 | 0.34 | 0.19 | 0.17 | 0.15 | 0.45 | Pcons_multi | 26 | |||||||||||||
| 27 | FFASflextemplate | 23.12 | 19.17 | 21.53 | 25.98 | 21.84 | 26.02 | 51.07 | 45.36 | 43.16 | 29.65 | 25.58 | 25.42 | 0.30 | 0.35 | 0.14 | 0.06 | 0.04 | 23.78 | 19.83 | 21.53 | 26.47 | 22.33 | 26.02 | 52.14 | 49.05 | 44.34 | 30.81 | 26.36 | 25.86 | 0.02 | FFASflextemplate | 27 | ||||||||||||||||||
| 28 | 3D−JIGSAW_AEP | 20.68 | 18.11 | 22.74 | 35.78 | 31.32 | 29.34 | 55.71 | 50.71 | 47.21 | 18.61 | 17.83 | 15.21 | 0.14 | 1.22 | 1.21 | 0.44 | 0.32 | 0.49 | 21.99 | 19.36 | 23.57 | 36.11 | 31.75 | 29.66 | 57.86 | 54.76 | 50.15 | 18.61 | 17.83 | 15.27 | 1.08 | 1.03 | 0.06 | 0.01 | 0.56 | 3D−JIGSAW_AEP | 28 | |||||||||||||
| 29 | Phyre2 | 20.39 | 16.32 | 23.32 | 13.72 | 11.93 | 20.66 | 61.07 | 49.88 | 56.62 | 38.37 | 34.50 | 37.61 | 0.90 | 0.39 | 1.01 | 0.99 | 1.03 | 0.23 | 21.71 | 17.26 | 24.69 | 16.01 | 15.20 | 21.59 | 65.00 | 55.24 | 60.26 | 39.53 | 37.60 | 39.43 | 0.95 | 0.64 | 1.04 | 0.73 | 1.00 | Phyre2 | 29 | |||||||||||||
| 30 | Phragment | 19.55 | 16.35 | 25.30 | 13.72 | 11.98 | 22.27 | 60.36 | 51.79 | 56.96 | 39.53 | 36.05 | 42.86 | 0.11 | 0.82 | 0.62 | 1.05 | 1.17 | 1.28 | 0.80 | 20.02 | 16.95 | 25.91 | 13.72 | 11.98 | 23.39 | 64.29 | 55.24 | 59.81 | 39.53 | 37.21 | 47.61 | 0.86 | 0.64 | 0.97 | 0.73 | 0.93 | 0.94 | Phragment | 30 | |||||||||||
| 31 | Poing | 19.55 | 15.73 | 28.93 | 13.56 | 10.62 | 26.93 | 62.14 | 50.95 | 56.59 | 42.44 | 38.95 | 47.99 | 0.57 | 0.15 | 1.01 | 0.51 | 1.00 | 1.63 | 1.74 | 1.36 | 20.02 | 17.14 | 28.93 | 21.08 | 19.01 | 30.62 | 62.14 | 51.19 | 56.59 | 42.44 | 38.95 | 47.99 | 0.27 | 0.20 | 0.57 | 0.01 | 0.50 | 1.25 | 1.22 | 0.99 | Poing | 31 | ||||||||
| 32 | circle | 19.45 | 16.51 | 15.19 | 20.75 | 17.81 | 11.35 | 50.71 | 47.38 | 42.44 | 33.72 | 27.91 | 41.49 | 0.09 | 0.26 | 0.65 | 25.94 | 19.42 | 22.35 | 32.35 | 25.71 | 29.97 | 59.64 | 56.67 | 52.55 | 39.53 | 36.05 | 44.88 | 0.31 | 0.59 | 0.19 | 0.11 | 0.24 | 0.86 | 0.73 | 0.74 | 0.60 | circle | 32 | ||||||||||||
| 33 | MULTICOM−RANK | 18.89 | 16.32 | 20.53 | 15.69 | 12.31 | 17.15 | 57.86 | 53.21 | 52.09 | 30.81 | 25.00 | 38.94 | 0.55 | 0.79 | 0.39 | 0.38 | 23.59 | 20.33 | 26.69 | 25.65 | 23.91 | 25.33 | 60.00 | 54.52 | 52.56 | 32.56 | 26.94 | 38.94 | 0.12 | 0.29 | 0.52 | MULTICOM−RANK | 33 | |||||||||||||||||
| 34 | MULTICOM−CMFR | 18.80 | 16.92 | 30.43 | 32.68 | 29.63 | 33.86 | 55.71 | 50.48 | 51.50 | 30.81 | 25.00 | 39.08 | 0.76 | 0.88 | 1.01 | 1.00 | 0.32 | 0.46 | 0.31 | 0.39 | 32.33 | 27.38 | 31.76 | 32.68 | 29.68 | 33.86 | 62.14 | 53.33 | 57.35 | 30.81 | 26.16 | 39.08 | 1.33 | 1.57 | 0.68 | 0.63 | 0.74 | 0.64 | 0.57 | 0.34 | 0.61 | MULTICOM−CMFR | 34 | |||||||
| 35 | MUProt | 18.70 | 16.82 | 30.45 | 32.84 | 29.68 | 33.78 | 56.07 | 50.12 | 51.81 | 30.81 | 27.13 | 39.02 | 0.77 | 0.90 | 1.01 | 0.99 | 0.36 | 0.42 | 0.36 | 0.39 | 32.14 | 25.94 | 34.50 | 33.99 | 29.68 | 34.67 | 67.50 | 54.52 | 65.44 | 31.98 | 27.13 | 39.02 | 1.30 | 1.27 | 1.08 | 0.80 | 0.74 | 0.75 | 1.28 | 0.52 | 1.80 | MUProt | 35 | |||||||
| 36 | panther_server | 18.61 | 16.16 | 12.92 | 14.71 | 12.74 | 10.28 | 55.00 | 51.67 | 46.81 | 27.33 | 24.61 | 26.19 | 0.25 | 0.60 | 19.08 | 16.26 | 15.52 | 16.67 | 15.63 | 11.28 | 55.71 | 51.67 | 49.13 | 36.05 | 29.46 | 34.65 | 0.08 | 0.11 | panther_server | 36 | ||||||||||||||||||||
| 37 | GeneSilicoMetaServer | 18.05 | 16.35 | 10.37 | 13.40 | 11.22 | 6.19 | 57.14 | 53.93 | 50.79 | 22.09 | 19.77 | 22.89 | 0.48 | 0.87 | 0.22 | 25.56 | 19.67 | 23.17 | 29.57 | 25.65 | 27.12 | 59.64 | 55.59 | 50.79 | 29.07 | 26.16 | 30.80 | 0.25 | 0.23 | 0.19 | 0.24 | 0.69 | GeneSilicoMetaServer | 37 | ||||||||||||||||
| 38 | FFASsuboptimal | 18.05 | 15.41 | 12.16 | 16.50 | 14.38 | 9.63 | 52.50 | 48.69 | 47.23 | 27.91 | 25.00 | 23.64 | 0.24 | 18.80 | 16.35 | 19.34 | 19.61 | 17.59 | 14.95 | 57.14 | 52.38 | 52.52 | 35.47 | 31.98 | 39.60 | 0.19 | 0.01 | 0.07 | FFASsuboptimal | 38 | ||||||||||||||||||||
| 39 | MUFOLD−MD | 17.76 | 13.94 | 41.40 | 31.21 | 23.91 | 44.68 | 46.79 | 37.98 | 53.62 | 45.93 | 44.38 | 61.36 | 2.16 | 0.72 | 0.31 | 2.33 | 0.60 | 2.18 | 2.60 | 2.82 | 17.76 | 13.94 | 41.40 | 31.21 | 23.91 | 44.68 | 61.43 | 51.43 | 56.61 | 55.81 | 49.23 | 61.36 | 2.08 | 0.44 | 2.13 | 0.48 | 0.05 | 0.50 | 3.64 | 2.92 | 2.69 | MUFOLD−MD | 39 | |||||||
| 40 | PSI | 17.57 | 14.91 | 25.61 | 20.10 | 17.05 | 23.68 | 53.21 | 47.50 | 47.71 | 40.12 | 36.24 | 52.49 | 0.15 | 0.05 | 0.10 | 1.27 | 1.31 | 1.85 | 17.57 | 14.91 | 27.95 | 20.10 | 17.05 | 26.63 | 53.57 | 47.86 | 49.32 | 50.58 | 47.87 | 60.00 | 0.13 | 2.70 | 2.70 | 2.52 | PSI | 40 | ||||||||||||||
| 41 | PS2−server | 17.57 | 14.25 | 17.61 | 16.34 | 15.63 | 16.51 | 51.79 | 47.14 | 46.94 | 26.16 | 24.23 | 30.77 | 0.06 | 26.13 | 20.96 | 26.71 | 19.44 | 17.05 | 28.21 | 64.29 | 56.07 | 59.96 | 34.30 | 28.10 | 38.60 | 0.34 | 0.25 | 0.86 | 0.76 | 0.99 | PS2−server | 41 | ||||||||||||||||||
| 42 | rehtnap | 16.45 | 13.94 | 3.41 | 14.54 | 12.36 | 4.34 | 39.64 | 38.33 | 23.00 | 26.16 | 22.29 | 22.03 | 16.92 | 14.72 | 6.29 | 14.71 | 12.96 | 4.79 | 42.86 | 39.29 | 35.58 | 26.16 | 22.87 | 22.03 | rehtnap | 42 | ||||||||||||||||||||||||
| 43 | Pcons_local | 15.51 | 14.25 | 0.77 | 10.62 | 9.31 | 55.71 | 50.95 | 50.34 | 0.00 | 0.00 | 5.09 | 0.32 | 0.51 | 0.16 | 15.51 | 14.25 | 0.77 | 10.62 | 9.31 | 55.71 | 50.95 | 50.34 | 0.00 | 0.00 | 5.09 | Pcons_local | 43 | |||||||||||||||||||||||
| 44 | SAM−T02−server | 15.32 | 11.31 | 7.84 | 7.19 | 53.57 | 50.48 | 50.82 | 6.98 | 6.98 | 5.67 | 0.09 | 0.46 | 0.22 | 15.70 | 14.07 | 7.53 | 24.18 | 23.53 | 7.72 | 55.71 | 52.50 | 53.22 | 20.35 | 18.80 | 15.49 | 0.21 | 0.00 | SAM−T02−server | 44 | |||||||||||||||||||||
| 45 | 3Dpro | 15.13 | 13.75 | 21.12 | 13.56 | 11.87 | 19.08 | 55.00 | 49.76 | 51.69 | 30.23 | 29.07 | 37.69 | 0.25 | 0.37 | 0.34 | 0.17 | 0.24 | 19.08 | 14.76 | 23.71 | 16.18 | 15.30 | 21.08 | 55.00 | 49.76 | 52.73 | 34.88 | 34.88 | 46.87 | 0.55 | 0.85 | 3Dpro | 45 | |||||||||||||||||
| 46 | GS−MetaServer2 | 15.04 | 11.22 | 2.61 | 2.61 | 53.57 | 44.29 | 50.56 | 18.02 | 13.76 | 14.10 | 0.09 | 0.19 | 25.56 | 19.36 | 23.17 | 27.94 | 23.80 | 27.12 | 59.64 | 55.59 | 50.79 | 28.49 | 26.16 | 30.80 | 0.25 | 0.02 | 0.24 | 0.69 | GS−MetaServer2 | 46 | ||||||||||||||||||||
| 47 | FOLDpro | 14.85 | 13.00 | 23.71 | 13.89 | 12.36 | 21.08 | 53.93 | 46.79 | 52.73 | 32.56 | 25.97 | 44.61 | 0.13 | 0.02 | 0.48 | 0.08 | 0.99 | 19.64 | 16.23 | 23.71 | 16.18 | 15.30 | 21.08 | 55.71 | 49.76 | 52.73 | 33.72 | 29.07 | 44.61 | 0.56 | FOLDpro | 47 | ||||||||||||||||||
| 48 | FALCON_CONSENSUS | 13.44 | 11.75 | 24.05 | 15.69 | 14.81 | 24.14 | 48.57 | 43.09 | 45.42 | 41.28 | 39.73 | 44.99 | 1.45 | 1.87 | 1.03 | 14.29 | 12.91 | 26.77 | 16.67 | 14.81 | 28.71 | 50.36 | 46.67 | 47.32 | 43.02 | 39.73 | 51.44 | 1.35 | 1.35 | 1.43 | FALCON_CONSENSUS | 48 | ||||||||||||||||||
| 49 | mariner1 | 13.16 | 11.72 | 14.63 | 14.71 | 11.87 | 18.49 | 50.36 | 44.88 | 50.58 | 0.00 | 0.00 | 5.09 | 0.19 | 17.39 | 14.54 | 21.01 | 20.75 | 14.43 | 23.97 | 66.43 | 55.83 | 62.93 | 34.88 | 31.59 | 44.83 | 1.14 | 0.73 | 1.43 | 0.00 | 0.59 | mariner1 | 49 | ||||||||||||||||||
| 50 | FALCON | 12.97 | 12.47 | 26.77 | 15.69 | 13.29 | 28.71 | 48.57 | 46.67 | 44.02 | 43.02 | 37.98 | 45.64 | 0.30 | 0.37 | 0.00 | 1.72 | 1.59 | 1.11 | 14.29 | 12.91 | 26.77 | 16.67 | 14.81 | 28.71 | 50.36 | 46.67 | 47.32 | 43.02 | 39.73 | 51.44 | 1.35 | 1.35 | 1.43 | FALCON | 50 | |||||||||||||||
| 51 | BioSerf | 12.41 | 9.27 | 18.47 | 15.36 | 9.86 | 18.96 | 43.93 | 32.98 | 42.74 | 33.14 | 23.64 | 32.32 | 0.17 | 12.41 | 9.27 | 18.47 | 15.36 | 9.86 | 18.96 | 43.93 | 32.98 | 42.74 | 33.14 | 23.64 | 32.32 | BioSerf | 51 | |||||||||||||||||||||||
| 52 | LOOPP_Server | 12.22 | 9.52 | 6.55 | 21.41 | 17.27 | 18.25 | 21.07 | 19.88 | 15.46 | 0.00 | 0.00 | 5.09 | 12.22 | 9.52 | 17.69 | 21.41 | 17.27 | 21.20 | 29.64 | 26.31 | 32.86 | 32.56 | 31.01 | 40.22 | 0.01 | LOOPP_Server | 52 | |||||||||||||||||||||||
| 53 | FUGUE_KM | 11.94 | 10.81 | 14.15 | 14.05 | 12.31 | 12.74 | 41.79 | 37.26 | 40.15 | 31.98 | 27.91 | 35.22 | 24.72 | 21.59 | 18.63 | 28.11 | 25.27 | 18.71 | 56.07 | 52.02 | 51.61 | 31.98 | 27.91 | 35.22 | 0.12 | 0.38 | 0.04 | 0.13 | 0.14 | FUGUE_KM | 53 | |||||||||||||||||||
| 54 | RBO−Proteus | 11.65 | 9.71 | 32.94 | 20.26 | 17.43 | 36.21 | 37.50 | 28.57 | 47.37 | 41.86 | 39.15 | 49.83 | 1.08 | 1.29 | 1.54 | 1.77 | 1.56 | 14.47 | 12.41 | 35.24 | 25.33 | 21.73 | 40.51 | 37.50 | 34.29 | 47.37 | 43.02 | 40.31 | 49.83 | 1.19 | 1.56 | 1.35 | 1.45 | 1.22 | RBO−Proteus | 54 | ||||||||||||||
| 55 | Pushchino | 10.81 | 8.11 | 11.50 | 18.63 | 14.05 | 25.18 | 31.07 | 27.74 | 13.77 | 19.77 | 15.50 | 9.95 | 10.81 | 8.11 | 11.50 | 18.63 | 14.05 | 25.18 | 31.07 | 27.74 | 13.77 | 19.77 | 15.50 | 9.95 | Pushchino | 55 | ||||||||||||||||||||||||
| 56 | FAMSD | 10.81 | 7.61 | 16.64 | 13.56 | 12.74 | 17.78 | 28.93 | 23.93 | 37.99 | 29.07 | 25.97 | 33.45 | 21.05 | 17.42 | 21.21 | 18.95 | 16.18 | 22.81 | 60.00 | 52.38 | 54.69 | 37.21 | 34.30 | 39.15 | 0.29 | 0.19 | 0.22 | 0.32 | 0.45 | FAMSD | 56 | |||||||||||||||||||
| 57 | forecast | 10.34 | 9.09 | 19.31 | 15.85 | 13.34 | 22.45 | 29.29 | 27.62 | 34.10 | 31.39 | 30.43 | 38.18 | 0.39 | 0.29 | 10.53 | 9.68 | 19.31 | 16.99 | 14.81 | 22.45 | 30.00 | 28.09 | 34.10 | 40.12 | 35.85 | 38.18 | 0.84 | 0.71 | forecast | 57 | ||||||||||||||||||||
| 58 | MUFOLD−Server | 10.24 | 7.86 | 19.54 | 15.03 | 8.99 | 20.45 | 31.79 | 24.76 | 38.60 | 36.05 | 18.41 | 35.22 | 0.63 | 10.43 | 8.40 | 23.60 | 16.18 | 12.42 | 26.07 | 32.14 | 25.00 | 40.26 | 38.37 | 25.19 | 35.91 | 0.52 | MUFOLD−Server | 58 | ||||||||||||||||||||||
| 59 | Distill | 9.96 | 8.99 | 19.90 | 17.16 | 15.30 | 22.70 | 26.07 | 23.93 | 35.50 | 38.95 | 34.30 | 39.13 | 1.08 | 1.00 | 0.40 | 11.00 | 8.99 | 23.32 | 18.95 | 15.30 | 28.12 | 28.93 | 25.00 | 36.35 | 38.95 | 34.69 | 41.27 | 0.63 | 0.52 | 0.14 | Distill | 59 | ||||||||||||||||||
| 60 | RANDOM | 9.02 | 7.79 | 9.02 | 12.92 | 10.78 | 12.92 | 29.31 | 23.55 | 29.31 | 30.40 | 27.26 | 30.40 | 9.02 | 7.79 | 9.02 | 12.92 | 10.78 | 12.92 | 29.31 | 23.55 | 29.31 | 30.40 | 27.26 | 30.40 | RANDOM | 60 | ||||||||||||||||||||||||
| 61 | LEE−SERVER | 9.02 | 7.33 | 14.50 | 11.77 | 10.73 | 15.43 | 26.07 | 25.59 | 30.95 | 43.02 | 36.43 | 39.46 | 1.72 | 1.34 | 0.43 | 13.16 | 11.03 | 26.15 | 21.89 | 16.78 | 33.75 | 33.93 | 27.26 | 40.54 | 43.02 | 36.43 | 39.46 | 0.63 | 1.35 | 0.80 | LEE−SERVER | 61 | ||||||||||||||||||
| 62 | huber−torda−server | 8.84 | 8.14 | 10.37 | 13.89 | 12.47 | 15.61 | 23.57 | 21.43 | 20.16 | 31.39 | 26.74 | 30.44 | 10.34 | 8.14 | 12.62 | 13.89 | 12.47 | 15.61 | 39.29 | 31.91 | 38.93 | 36.63 | 35.08 | 35.16 | 0.21 | 0.58 | huber−torda−server | 62 | ||||||||||||||||||||||
| 63 | OLGAFS | 8.74 | 7.74 | 2.14 | 12.09 | 10.95 | 3.19 | 25.71 | 21.43 | 21.11 | 24.42 | 20.93 | 25.13 | 14.10 | 11.47 | 12.07 | 12.09 | 10.95 | 10.23 | 51.07 | 47.02 | 43.17 | 27.33 | 23.84 | 28.40 | OLGAFS | 63 | ||||||||||||||||||||||||
| 64 | schenk−torda−server | 8.55 | 8.27 | 13.14 | 11.60 | 11.60 | 15.15 | 27.50 | 26.55 | 32.87 | 33.72 | 30.62 | 30.78 | 0.26 | 0.42 | 8.55 | 8.27 | 13.14 | 11.60 | 11.60 | 15.15 | 27.50 | 26.55 | 32.87 | 34.88 | 30.62 | 37.34 | schenk−torda−server | 64 | ||||||||||||||||||||||
| 65 | SAM−T06−server | 8.18 | 7.24 | 14.68 | 14.38 | 12.64 | 14.98 | 25.36 | 22.98 | 27.77 | 39.53 | 38.37 | 46.70 | 1.17 | 1.65 | 1.22 | 24.44 | 20.24 | 16.31 | 30.07 | 28.76 | 20.54 | 52.14 | 49.52 | 50.10 | 39.53 | 38.37 | 46.70 | 0.07 | 0.10 | 0.29 | 0.62 | 0.73 | 1.12 | 0.83 | SAM−T06−server | 65 | ||||||||||||||
| 66 | pipe_int | 7.89 | 6.36 | 15.80 | 11.93 | 10.68 | 15.26 | 21.07 | 17.14 | 36.44 | 26.74 | 25.97 | 40.02 | 0.49 | 7.89 | 6.36 | 15.80 | 11.93 | 10.68 | 15.26 | 21.07 | 17.14 | 36.44 | 26.74 | 25.97 | 40.02 | pipe_int | 66 | |||||||||||||||||||||||
| 67 | ACOMPMOD | ACOMPMOD | 67 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 68 | BHAGEERATH | BHAGEERATH | 68 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 69 | Fiser−M4T | Fiser−M4T | 69 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 70 | Frankenstein | Frankenstein | 70 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 71 | YASARA | YASARA | 71 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 72 | keasar−server | keasar−server | 72 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 73 | mahmood−torda−server | mahmood−torda−server | 73 | ||||||||||||||||||||||||||||||||||||||||||||||||
| 74 | test_http_server_01 | test_http_server_01 | 74 | ||||||||||||||||||||||||||||||||||||||||||||||||