T0422
fidgetin-like protein 1 (FIGNL1A) from Homo sapiens
Target sequence:
>T0422 FIGNL1A, Homo sapiens, 357 residues
MHHHHHHSSGVDLGTENLYFQSMVPPIPKQDGGEQNGGMQCKPYGAGPTEPAHPVDERLKNLEPKMIELIMNEIMDHGPPVNWEDIAGVEFAKATIKEIVVWPMLRPDIFTGLRGPPKGILLFGPPGTGKTLIGKCIASQSGATFFSISASSLTSKWVGEGEKMVRALFAVARCQQPAVIFIDEIDSLLSQRGDGEHESSRRIKTEFLVQLDGATTSSEDRILVVGATNRPQEIDEAARRRLVKRLYIPLPEASARKQIVINLMSKEQCCLSEEEIEQIVQQSDAFSGADMTQLCREASLGPIRSLQTADIATITPDQVRPIAYIDFENAFRTVRPSVSPKDLELYENWNKTFGCGK
Structure:
Determined by:
SGC
PDB ID: 3d8b
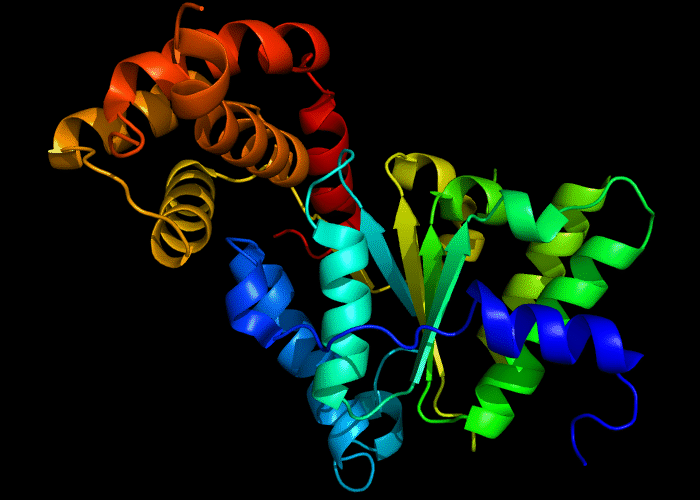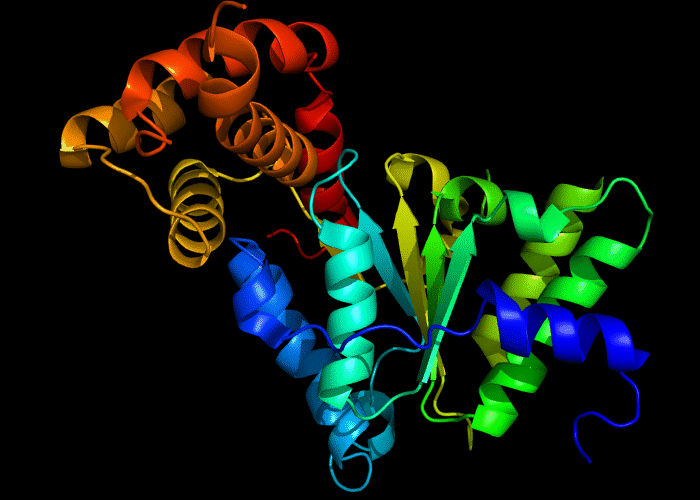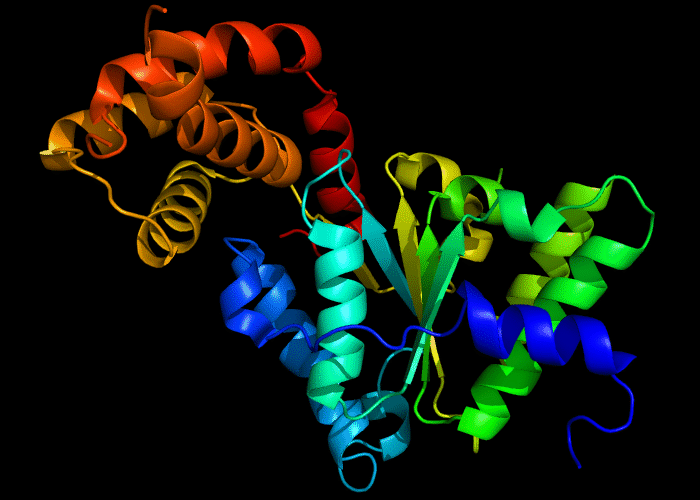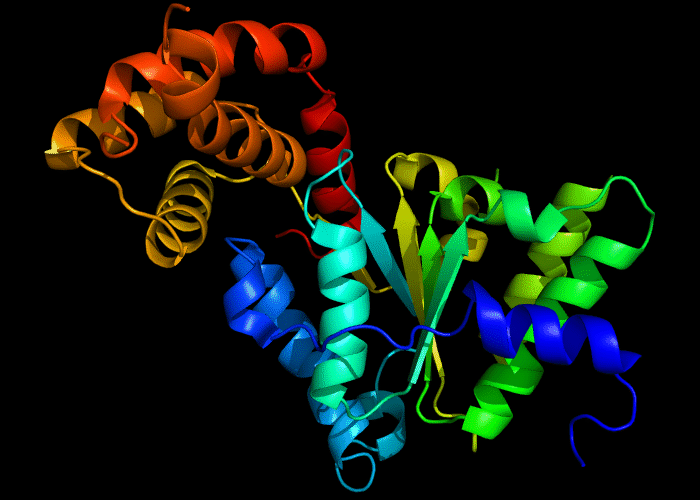
Cartoon diagram of 422: 3d8b chain A
Domains: PyMOL of domains
Two domains. Residue ranges in PDB: 374-567,657-674 and 568-656. Residue ranges in target: 57-250,340-357 and 251-339.
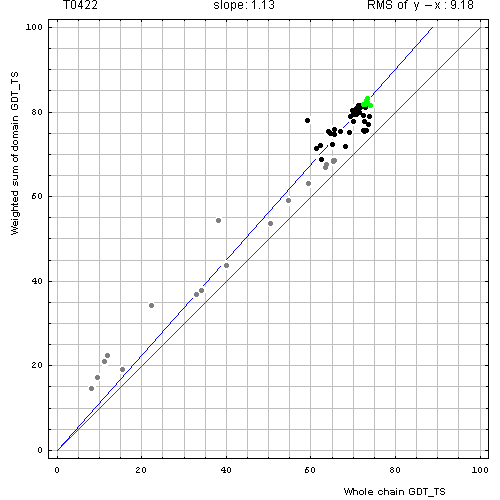
Correlation between weighted by the number of residues sum of GDT-TS scores for domain-based evlatuation (y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
To compute the weighted sum, GDT-TS for each domain was multiplied by the domain length, and this sum was divided by the sum of domain lengths. Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the weighted sum of domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
N-domain: AAA+ ATPase from P-loop-containing nucleoside triphosphate hydrolase fold. C-domain: Histone fold.
CASP category:
Whole chain: Comparative modeling:medium.
1st domain: Comparative modeling:medium. 2nd domain: Comparative modeling:medium. As the categories are the same for the whole chain and domains, domain partitioning may not be necessary for evaluation.
Closest templates:
1ypw, 1e32, 3cf0, 2qp9, 1xwi, 3b9p.
Target sequence - PDB file inconsistencies:
T0422 3d8b.pdb T0422.pdb PyMOL PyMOL of domains
T0422 1 MHHHHHHSSGVDLGTENLYFQSMVPPIPKQDGGEQNGGMQCKPYGAGPTEPAHPVDERLKNLEPKMIELIMNEIMDHGPPVNWEDIAGVEFAKATIKEIVVWPMLRPDIFTGLRGPPKGILLFGPPGTGKTLIGKCIASQSGATFFSISASSLTSKWVGEGEKMVRALFAVARCQQPAVIFIDEIDSLLSQRGDGEHESSRRIKTEFLVQLDGATTSSEDRILVVGATNRPQEIDEAARRRLVKRLYIPLPEASARKQIVINLMSKEQCCLSEEEIEQIVQQSDAFSGADMTQLCREASLGPIRSLQTADIATITPDQVRPIAYIDFENAFRTVRPSVSPKDLELYENWNKTFGCGK 357 ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~||||||||||||||||~~~~|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~||||||||||||||||||||||||||||||||||||||| 3d8bA 374 --------------------------------------------------------ERLKNLEPKMIELIMNEIMDHGPPVNWEDIAGVEFAKATIKEIVVWPMLRPDIFTGLRGPPKGILLFGPPGTGKTLIGKCIASQSGATFFSISASSLTSKWVGEGEKMVRALFAVARCQQPAVIFIDEIDSLLSQ------ESSRRIKTEFLVQLDG----SEDRILVVGATNRPQEIDEAARRRLVKRLYIPLPEASARKQIVINLMSKEQCCLSEEEIEQIVQQSDAFSGADMTQLCREASLGPIRSLQT----------VRPIAYIDFENAFRTVRPSVSPKDLELYENWNKTFGCGK 674
We suggest to evaluate this target as a single domain spanning whole chain. However, evolutionarily this is a 2-domain protein:
1st domain: target 57-191, 198-213, 218-250, 340-357 ; pdb 374-508, 515-530, 535-567, 657-674
2nd domain: target 251-308, 319-339 ; pdb 568-625, 636-656
Sequence classification:
N-domain: ATPase family associated with various cellular activities (AAA), C-domain: Vps4 C terminal oligomerisation domain (Vps4_C) in Pfam.
Server predictions:
T0422:pdb 374-674:seq 57-357:CM_medium; alignment
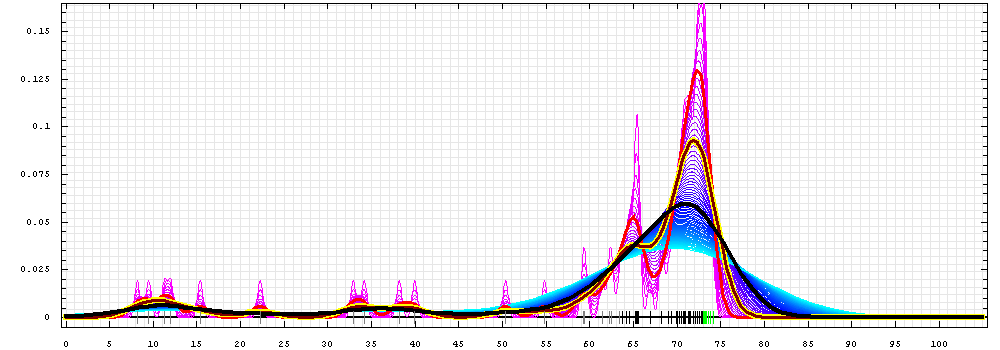
First models for T0422:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
422_1:pdb 374-567,657-674:seq 57-250,340-357:CM_medium; alignment
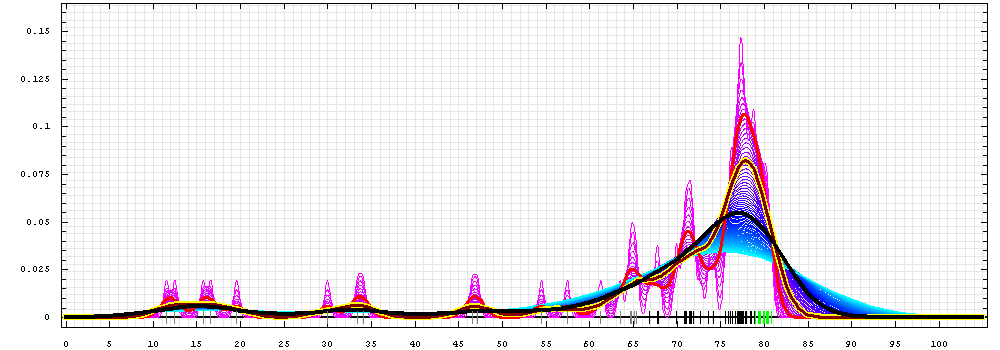
First models for T0422_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
422_2:pdb 568-656:seq 251-339:CM_easy; alignment
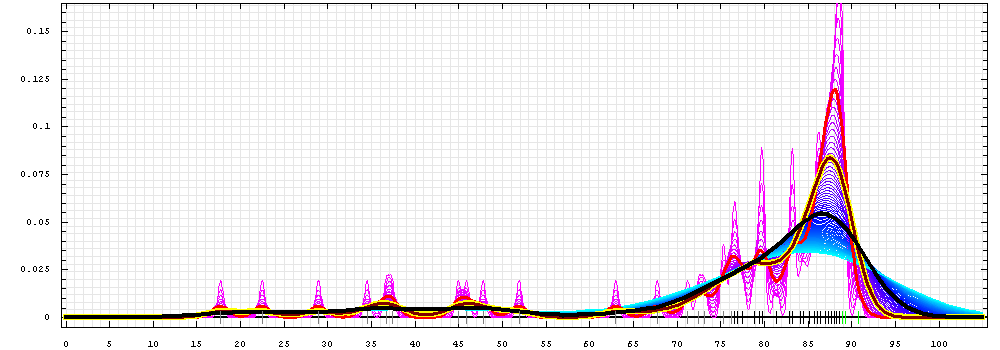
First models for T0422_2:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0422 | 422_1 | 422_2 | T0422 | 422_1 | 422_2 | T0422 | 422_1 | 422_2 | T0422 | 422_1 | 422_2 | ||||||||||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||||||||||||||
| 1 | 3D−JIGSAW_AEP | 74.11 | 67.59 | 79.51 | 79.95 | 75.58 | 79.26 | 85.76 | 81.75 | 84.26 | 0.89 | 0.94 | 0.90 | 0.92 | 0.88 | 1.00 | 0.43 | 0.52 | 0.70 | 74.47 | 68.42 | 79.67 | 81.31 | 77.93 | 79.76 | 86.71 | 83.75 | 84.42 | 1.07 | 1.14 | 0.97 | 1.11 | 1.16 | 1.14 | 0.41 | 0.55 | 0.63 | 3D−JIGSAW_AEP | 1 |
| 2 | MULTICOM−CLUSTER | 73.84 | 69.04 | 75.44 | 77.35 | 73.89 | 75.44 | 82.91 | 68.56 | 79.77 | 0.85 | 1.14 | 0.47 | 0.57 | 0.69 | 0.55 | 0.09 | 0.18 | 73.84 | 69.75 | 76.22 | 77.47 | 73.89 | 75.44 | 86.08 | 74.16 | 82.62 | 0.92 | 1.46 | 0.41 | 0.35 | 0.48 | 0.37 | 0.29 | 0.34 | MULTICOM−CLUSTER | 2 | ||
| 3 | MULTICOM−RANK | 73.58 | 68.24 | 73.58 | 75.87 | 67.95 | 74.83 | 79.75 | 69.41 | 75.36 | 0.81 | 1.03 | 0.28 | 0.38 | 0.48 | 73.58 | 68.68 | 73.58 | 75.87 | 67.95 | 74.83 | 80.38 | 74.16 | 76.09 | 0.86 | 1.20 | 0.04 | 0.26 | MULTICOM−RANK | 3 | |||||||||
| 4 | METATASSER | 73.31 | 66.55 | 78.66 | 79.83 | 77.27 | 78.39 | 88.29 | 85.34 | 84.30 | 0.78 | 0.80 | 0.81 | 0.90 | 1.08 | 0.89 | 0.73 | 0.87 | 0.70 | 74.91 | 71.47 | 78.66 | 80.32 | 78.38 | 78.39 | 88.29 | 85.34 | 84.30 | 1.17 | 1.88 | 0.80 | 0.91 | 1.23 | 0.89 | 0.69 | 0.79 | 0.61 | METATASSER | 4 |
| 5 | BAKER−ROBETTA | 73.31 | 65.95 | 80.63 | 80.82 | 74.96 | 81.19 | 88.92 | 85.55 | 83.43 | 0.78 | 0.72 | 1.01 | 1.03 | 0.81 | 1.22 | 0.81 | 0.89 | 0.60 | 74.38 | 67.97 | 81.33 | 81.81 | 77.19 | 82.23 | 88.92 | 87.45 | 83.98 | 1.05 | 1.03 | 1.23 | 1.21 | 1.03 | 1.58 | 0.81 | 1.12 | 0.56 | BAKER−ROBETTA | 5 |
| 6 | COMA−M | 73.22 | 64.68 | 78.56 | 80.20 | 78.30 | 77.97 | 88.61 | 77.22 | 84.50 | 0.76 | 0.55 | 0.80 | 0.95 | 1.20 | 0.85 | 0.77 | 0.07 | 0.73 | 73.22 | 64.68 | 79.55 | 80.82 | 78.67 | 78.99 | 88.61 | 85.65 | 85.00 | 0.77 | 0.24 | 0.95 | 1.01 | 1.28 | 1.00 | 0.75 | 0.84 | 0.72 | COMA−M | 6 |
| 7 | MULTICOM−REFINE | 73.13 | 68.98 | 72.69 | 74.13 | 67.37 | 73.35 | 79.43 | 77.74 | 76.07 | 0.75 | 1.13 | 0.18 | 0.15 | 0.31 | 0.12 | 73.84 | 70.34 | 75.14 | 76.86 | 70.67 | 74.93 | 82.91 | 77.74 | 80.21 | 0.92 | 1.60 | 0.24 | 0.23 | 0.28 | MULTICOM−REFINE | 7 | |||||||
| 8 | keasar−server | 73.04 | 66.40 | 78.59 | 80.45 | 75.50 | 78.07 | 88.61 | 87.13 | 84.48 | 0.74 | 0.78 | 0.80 | 0.98 | 0.87 | 0.86 | 0.77 | 1.04 | 0.72 | 73.04 | 66.40 | 78.59 | 80.45 | 75.50 | 78.08 | 89.24 | 87.76 | 84.48 | 0.73 | 0.66 | 0.79 | 0.94 | 0.75 | 0.84 | 0.86 | 1.16 | 0.64 | keasar−server | 8 |
| 9 | GS−KudlatyPred | 73.04 | 64.44 | 76.67 | 80.32 | 76.03 | 76.07 | 87.34 | 85.23 | 83.28 | 0.74 | 0.52 | 0.60 | 0.96 | 0.94 | 0.63 | 0.62 | 0.86 | 0.59 | 73.04 | 64.44 | 76.67 | 80.32 | 76.03 | 76.07 | 87.34 | 85.23 | 83.28 | 0.73 | 0.19 | 0.48 | 0.91 | 0.84 | 0.48 | 0.52 | 0.78 | 0.44 | GS−KudlatyPred | 9 |
| 10 | SAM−T08−server | 72.86 | 67.47 | 77.95 | 78.47 | 75.25 | 76.97 | 87.34 | 80.38 | 84.73 | 0.71 | 0.93 | 0.73 | 0.72 | 0.84 | 0.73 | 0.62 | 0.38 | 0.75 | 72.86 | 67.47 | 78.39 | 78.47 | 75.25 | 78.05 | 88.61 | 80.38 | 84.73 | 0.69 | 0.91 | 0.76 | 0.55 | 0.71 | 0.83 | 0.75 | 0.04 | 0.68 | SAM−T08−server | 10 |
| 11 | YASARA | 72.86 | 66.16 | 80.70 | 78.96 | 76.07 | 79.97 | 88.61 | 85.02 | 85.93 | 0.71 | 0.75 | 1.02 | 0.79 | 0.94 | 1.08 | 0.77 | 0.84 | 0.89 | 72.86 | 66.16 | 80.70 | 78.96 | 76.07 | 79.97 | 88.61 | 85.55 | 86.18 | 0.69 | 0.60 | 1.13 | 0.65 | 0.85 | 1.18 | 0.75 | 0.83 | 0.91 | YASARA | 11 |
| 12 | PSI | 72.86 | 65.98 | 79.36 | 79.58 | 74.22 | 79.10 | 88.61 | 80.38 | 84.68 | 0.71 | 0.72 | 0.88 | 0.87 | 0.73 | 0.98 | 0.77 | 0.38 | 0.75 | 72.86 | 66.40 | 79.50 | 79.58 | 75.12 | 79.29 | 88.61 | 80.80 | 84.68 | 0.69 | 0.66 | 0.94 | 0.77 | 0.69 | 1.06 | 0.75 | 0.10 | 0.67 | PSI | 12 |
| 13 | MUProt | 72.78 | 62.10 | 72.46 | 73.64 | 70.26 | 73.08 | 79.75 | 70.67 | 75.99 | 0.70 | 0.20 | 0.16 | 0.09 | 0.27 | 0.28 | 72.78 | 68.06 | 72.46 | 73.89 | 70.42 | 73.08 | 80.38 | 78.38 | 76.11 | 0.67 | 1.06 | MUProt | 13 | ||||||||||
| 14 | pro−sp3−TASSER | 72.60 | 64.41 | 73.41 | 76.36 | 71.41 | 73.77 | 81.33 | 74.37 | 77.01 | 0.68 | 0.51 | 0.26 | 0.44 | 0.40 | 0.36 | 74.20 | 68.21 | 79.82 | 81.19 | 79.45 | 79.86 | 88.92 | 77.32 | 84.53 | 1.00 | 1.09 | 0.99 | 1.08 | 1.41 | 1.16 | 0.81 | 0.64 | pro−sp3−TASSER | 14 | ||||
| 15 | MULTICOM−CMFR | 72.51 | 63.97 | 72.13 | 74.13 | 66.09 | 72.67 | 79.75 | 77.22 | 75.90 | 0.67 | 0.45 | 0.12 | 0.15 | 0.23 | 0.07 | 72.51 | 63.97 | 72.13 | 74.13 | 67.12 | 72.71 | 80.06 | 77.22 | 78.52 | 0.60 | 0.07 | MULTICOM−CMFR | 15 | ||||||||||
| 16 | Zhang−Server | 72.33 | 66.10 | 79.24 | 79.45 | 74.50 | 79.48 | 86.08 | 77.00 | 82.44 | 0.64 | 0.74 | 0.87 | 0.85 | 0.76 | 1.02 | 0.47 | 0.05 | 0.49 | 73.93 | 69.36 | 79.58 | 81.06 | 79.00 | 79.65 | 87.66 | 81.65 | 83.71 | 0.94 | 1.37 | 0.95 | 1.06 | 1.34 | 1.12 | 0.58 | 0.23 | 0.51 | Zhang−Server | 16 |
| 17 | RAPTOR | 72.33 | 64.03 | 75.54 | 76.86 | 75.37 | 74.49 | 85.13 | 80.70 | 82.53 | 0.64 | 0.46 | 0.48 | 0.51 | 0.86 | 0.44 | 0.36 | 0.41 | 0.50 | 72.33 | 65.48 | 75.54 | 77.97 | 75.37 | 75.93 | 85.76 | 82.38 | 83.03 | 0.56 | 0.44 | 0.30 | 0.45 | 0.73 | 0.46 | 0.24 | 0.34 | 0.40 | RAPTOR | 17 |
| 18 | FFASstandard | 72.33 | 58.30 | 76.17 | 78.84 | 76.36 | 74.89 | 88.92 | 85.97 | 84.67 | 0.64 | 0.55 | 0.77 | 0.97 | 0.49 | 0.81 | 0.93 | 0.75 | 72.33 | 64.71 | 76.17 | 78.84 | 76.36 | 74.89 | 88.92 | 87.03 | 84.67 | 0.56 | 0.25 | 0.40 | 0.62 | 0.89 | 0.27 | 0.81 | 1.05 | 0.67 | FFASstandard | 18 | |
| 19 | FFASflextemplate | 72.33 | 58.30 | 76.17 | 78.84 | 76.36 | 74.89 | 88.92 | 85.97 | 84.67 | 0.64 | 0.55 | 0.77 | 0.97 | 0.49 | 0.81 | 0.93 | 0.75 | 72.33 | 65.18 | 76.17 | 78.84 | 76.36 | 74.89 | 88.92 | 87.03 | 84.67 | 0.56 | 0.36 | 0.40 | 0.62 | 0.89 | 0.27 | 0.81 | 1.05 | 0.67 | FFASflextemplate | 19 | |
| 20 | FAMSD | 72.15 | 65.15 | 77.81 | 78.47 | 75.74 | 77.21 | 89.24 | 86.08 | 84.20 | 0.62 | 0.61 | 0.72 | 0.72 | 0.90 | 0.76 | 0.84 | 0.94 | 0.69 | 72.15 | 65.36 | 77.81 | 78.47 | 75.74 | 77.21 | 89.24 | 86.08 | 84.21 | 0.52 | 0.41 | 0.67 | 0.55 | 0.79 | 0.68 | 0.86 | 0.91 | 0.59 | FAMSD | 20 |
| 21 | circle | 71.89 | 64.89 | 75.45 | 77.97 | 72.61 | 74.03 | 88.92 | 85.97 | 84.27 | 0.58 | 0.58 | 0.47 | 0.66 | 0.54 | 0.39 | 0.81 | 0.93 | 0.70 | 73.04 | 66.52 | 77.78 | 79.70 | 73.68 | 77.14 | 88.92 | 85.97 | 84.27 | 0.73 | 0.69 | 0.66 | 0.79 | 0.45 | 0.67 | 0.81 | 0.89 | 0.60 | circle | 21 |
| 22 | COMA | 71.89 | 64.29 | 77.35 | 78.84 | 75.78 | 76.58 | 87.97 | 86.50 | 84.28 | 0.58 | 0.50 | 0.67 | 0.77 | 0.91 | 0.69 | 0.69 | 0.98 | 0.70 | 71.89 | 64.29 | 79.55 | 78.84 | 75.78 | 78.99 | 87.97 | 86.50 | 85.00 | 0.46 | 0.15 | 0.95 | 0.62 | 0.80 | 1.00 | 0.64 | 0.97 | 0.72 | COMA | 22 |
| 23 | FFASsuboptimal | 71.53 | 64.71 | 75.84 | 77.85 | 75.54 | 74.58 | 88.29 | 87.03 | 84.05 | 0.53 | 0.55 | 0.51 | 0.64 | 0.88 | 0.45 | 0.73 | 1.03 | 0.67 | 71.53 | 64.71 | 75.84 | 77.85 | 75.54 | 74.58 | 88.29 | 87.03 | 84.05 | 0.37 | 0.25 | 0.35 | 0.43 | 0.76 | 0.22 | 0.69 | 1.05 | 0.57 | FFASsuboptimal | 23 |
| 24 | 3D−JIGSAW_V3 | 71.44 | 65.45 | 76.09 | 77.23 | 73.60 | 75.47 | 86.39 | 80.27 | 81.92 | 0.52 | 0.65 | 0.54 | 0.56 | 0.65 | 0.56 | 0.51 | 0.37 | 0.43 | 73.58 | 67.41 | 79.92 | 80.57 | 77.60 | 79.91 | 86.71 | 82.81 | 84.40 | 0.86 | 0.90 | 1.01 | 0.96 | 1.10 | 1.17 | 0.41 | 0.41 | 0.62 | 3D−JIGSAW_V3 | 24 |
| 25 | HHpred4 | 71.44 | 63.55 | 78.02 | 77.97 | 72.61 | 77.41 | 87.97 | 86.08 | 84.48 | 0.52 | 0.40 | 0.74 | 0.66 | 0.54 | 0.78 | 0.69 | 0.94 | 0.72 | 71.44 | 63.55 | 78.02 | 77.97 | 72.61 | 77.41 | 87.97 | 86.08 | 84.48 | 0.35 | 0.70 | 0.45 | 0.27 | 0.72 | 0.64 | 0.91 | 0.64 | HHpred4 | 25 | |
| 26 | Phyre_de_novo | 71.26 | 64.68 | 78.80 | 76.98 | 71.12 | 76.99 | 90.82 | 84.07 | 87.49 | 0.49 | 0.55 | 0.82 | 0.53 | 0.37 | 0.73 | 1.03 | 0.74 | 1.07 | 72.69 | 65.27 | 79.65 | 79.45 | 71.12 | 78.26 | 90.82 | 84.07 | 87.49 | 0.65 | 0.39 | 0.96 | 0.74 | 0.02 | 0.87 | 1.15 | 0.60 | 1.12 | Phyre_de_novo | 26 |
| 27 | MUSTER | 71.26 | 64.68 | 78.11 | 79.33 | 77.10 | 77.46 | 87.03 | 84.07 | 84.12 | 0.49 | 0.55 | 0.75 | 0.83 | 1.06 | 0.79 | 0.58 | 0.74 | 0.68 | 71.26 | 64.68 | 78.11 | 79.33 | 77.10 | 77.46 | 87.03 | 84.07 | 84.12 | 0.31 | 0.24 | 0.72 | 0.72 | 1.02 | 0.73 | 0.47 | 0.60 | 0.58 | MUSTER | 27 |
| 28 | LEE−SERVER | 70.91 | 64.38 | 78.78 | 78.34 | 73.14 | 77.51 | 88.29 | 80.06 | 85.91 | 0.44 | 0.51 | 0.82 | 0.70 | 0.60 | 0.79 | 0.73 | 0.35 | 0.89 | 72.15 | 65.92 | 78.84 | 79.33 | 73.14 | 77.67 | 89.87 | 85.55 | 86.60 | 0.52 | 0.54 | 0.83 | 0.72 | 0.36 | 0.77 | 0.98 | 0.83 | 0.97 | LEE−SERVER | 28 |
| 29 | forecast | 70.82 | 64.41 | 75.57 | 76.11 | 71.74 | 74.20 | 87.97 | 78.27 | 84.32 | 0.43 | 0.51 | 0.48 | 0.41 | 0.44 | 0.41 | 0.69 | 0.18 | 0.71 | 70.82 | 65.69 | 75.75 | 76.24 | 72.24 | 74.40 | 88.29 | 87.03 | 84.38 | 0.20 | 0.49 | 0.34 | 0.11 | 0.20 | 0.18 | 0.69 | 1.05 | 0.62 | forecast | 29 |
| 30 | Pushchino | 70.82 | 64.35 | 74.99 | 77.10 | 72.40 | 73.60 | 87.66 | 84.70 | 83.67 | 0.43 | 0.50 | 0.42 | 0.54 | 0.51 | 0.34 | 0.66 | 0.81 | 0.63 | 70.82 | 64.35 | 74.99 | 77.10 | 72.40 | 73.60 | 87.66 | 84.70 | 83.67 | 0.20 | 0.16 | 0.21 | 0.28 | 0.23 | 0.04 | 0.58 | 0.70 | 0.51 | Pushchino | 30 |
| 31 | panther_server | 70.73 | 64.27 | 74.09 | 76.36 | 71.99 | 72.85 | 88.61 | 86.08 | 81.68 | 0.42 | 0.49 | 0.33 | 0.44 | 0.47 | 0.26 | 0.77 | 0.94 | 0.40 | 70.73 | 64.27 | 74.09 | 76.36 | 71.99 | 72.85 | 88.61 | 86.08 | 81.68 | 0.18 | 0.14 | 0.07 | 0.13 | 0.16 | 0.75 | 0.91 | 0.19 | panther_server | 31 | |
| 32 | PS2−server | 70.55 | 60.05 | 75.47 | 77.60 | 73.23 | 74.25 | 87.34 | 83.75 | 83.84 | 0.39 | 0.47 | 0.61 | 0.61 | 0.42 | 0.62 | 0.71 | 0.65 | 70.55 | 60.35 | 75.47 | 77.60 | 73.23 | 74.25 | 87.34 | 83.75 | 83.84 | 0.14 | 0.29 | 0.38 | 0.37 | 0.16 | 0.52 | 0.55 | 0.53 | PS2−server | 32 | ||
| 33 | mGenTHREADER | 70.28 | 63.94 | 73.58 | 76.73 | 71.95 | 72.40 | 86.08 | 77.22 | 81.83 | 0.36 | 0.45 | 0.28 | 0.49 | 0.46 | 0.20 | 0.47 | 0.07 | 0.42 | 70.28 | 63.94 | 73.58 | 76.73 | 71.95 | 72.40 | 86.08 | 77.22 | 81.83 | 0.07 | 0.07 | 0.21 | 0.16 | 0.29 | 0.21 | mGenTHREADER | 33 | |||
| 34 | CpHModels | 70.02 | 64.56 | 71.82 | 77.35 | 73.72 | 74.12 | 78.80 | 75.00 | 70.46 | 0.32 | 0.53 | 0.09 | 0.57 | 0.67 | 0.40 | 70.02 | 64.56 | 71.82 | 77.35 | 73.72 | 74.12 | 78.80 | 75.00 | 70.46 | 0.01 | 0.21 | 0.33 | 0.45 | 0.13 | CpHModels | 34 | |||||||
| 35 | GeneSilicoMetaServer | 70.02 | 62.37 | 75.44 | 76.11 | 70.75 | 74.35 | 87.03 | 83.86 | 83.46 | 0.32 | 0.24 | 0.47 | 0.41 | 0.32 | 0.43 | 0.58 | 0.72 | 0.61 | 70.02 | 62.37 | 75.44 | 76.11 | 70.75 | 74.35 | 87.03 | 83.86 | 83.46 | 0.01 | 0.29 | 0.09 | 0.18 | 0.47 | 0.57 | 0.47 | GeneSilicoMetaServer | 35 | ||
| 36 | Fiser−M4T | 69.93 | 61.51 | 76.04 | 77.35 | 74.05 | 74.65 | 87.97 | 86.92 | 84.27 | 0.31 | 0.12 | 0.53 | 0.57 | 0.71 | 0.46 | 0.69 | 1.02 | 0.70 | 69.93 | 61.51 | 76.04 | 77.35 | 74.05 | 74.65 | 87.97 | 86.92 | 84.27 | 0.38 | 0.33 | 0.51 | 0.23 | 0.64 | 1.04 | 0.60 | Fiser−M4T | 36 | ||
| 37 | GS−MetaServer2 | 69.39 | 61.33 | 75.88 | 75.50 | 70.22 | 74.89 | 87.03 | 84.28 | 83.55 | 0.23 | 0.10 | 0.52 | 0.33 | 0.26 | 0.49 | 0.58 | 0.77 | 0.62 | 69.39 | 61.33 | 75.88 | 75.50 | 70.22 | 74.89 | 87.03 | 84.28 | 83.55 | 0.36 | 0.27 | 0.47 | 0.63 | 0.49 | GS−MetaServer2 | 37 | ||||
| 38 | Pcons_multi | 69.04 | 59.73 | 73.84 | 70.79 | 60.31 | 71.97 | 86.39 | 80.70 | 84.48 | 0.19 | 0.30 | 0.15 | 0.51 | 0.41 | 0.72 | 69.04 | 59.73 | 73.84 | 70.79 | 60.31 | 71.97 | 86.39 | 80.70 | 84.48 | 0.03 | 0.35 | 0.08 | 0.64 | Pcons_multi | 38 | ||||||||
| 39 | FALCON_CONSENSUS | 68.24 | 64.09 | 68.27 | 69.93 | 66.13 | 69.02 | 76.58 | 72.15 | 75.27 | 0.07 | 0.47 | 68.86 | 64.09 | 68.31 | 70.30 | 66.13 | 69.23 | 76.58 | 74.89 | 75.27 | 0.10 | FALCON_CONSENSUS | 39 | |||||||||||||||
| 40 | FALCON | 68.24 | 64.09 | 68.27 | 69.93 | 66.13 | 69.02 | 76.58 | 72.15 | 75.27 | 0.07 | 0.47 | 68.86 | 64.09 | 68.31 | 70.30 | 66.13 | 69.23 | 76.58 | 74.89 | 75.27 | 0.10 | FALCON | 40 | |||||||||||||||
| 41 | fais−server | 66.90 | 60.80 | 72.29 | 71.04 | 63.04 | 71.32 | 86.39 | 83.02 | 82.95 | 0.02 | 0.14 | 0.08 | 0.51 | 0.64 | 0.55 | 71.89 | 65.66 | 77.79 | 78.71 | 73.10 | 77.08 | 88.29 | 85.55 | 84.37 | 0.46 | 0.48 | 0.66 | 0.60 | 0.35 | 0.66 | 0.69 | 0.83 | 0.62 | fais−server | 41 | |||
| 42 | pipe_int | 65.57 | 60.11 | 71.57 | 70.92 | 68.44 | 71.11 | 84.18 | 79.96 | 81.01 | 0.06 | 0.06 | 0.05 | 0.24 | 0.34 | 0.33 | 71.80 | 65.45 | 77.97 | 78.96 | 74.50 | 77.51 | 87.97 | 85.23 | 83.80 | 0.43 | 0.43 | 0.69 | 0.65 | 0.58 | 0.74 | 0.64 | 0.78 | 0.53 | pipe_int | 42 | |||
| 43 | nFOLD3 | 65.48 | 59.96 | 71.70 | 72.65 | 62.58 | 69.79 | 84.49 | 74.37 | 81.81 | 0.08 | 0.28 | 0.42 | 65.48 | 60.53 | 71.70 | 72.65 | 62.58 | 69.79 | 84.49 | 76.58 | 81.81 | 0.01 | 0.21 | nFOLD3 | 43 | |||||||||||||
| 44 | 3Dpro | 65.48 | 59.02 | 61.75 | 65.35 | 59.74 | 61.17 | 76.27 | 69.73 | 70.66 | 65.48 | 59.02 | 61.75 | 65.35 | 59.74 | 61.64 | 76.27 | 72.36 | 70.66 | 3Dpro | 44 | ||||||||||||||||||
| 45 | ACOMPMOD | 65.30 | 60.44 | 60.57 | 64.73 | 57.88 | 59.40 | 77.53 | 76.90 | 71.97 | 0.04 | 65.30 | 60.44 | 60.57 | 68.32 | 60.64 | 60.57 | 77.53 | 76.90 | 71.97 | ACOMPMOD | 45 | |||||||||||||||||
| 46 | Pcons_dot_net | 65.30 | 59.02 | 61.66 | 65.10 | 60.31 | 60.90 | 76.90 | 74.58 | 71.81 | 65.30 | 60.50 | 61.66 | 69.68 | 67.04 | 61.88 | 77.22 | 75.95 | 71.81 | Pcons_dot_net | 46 | ||||||||||||||||||
| 47 | 3DShot2 | 65.21 | 58.93 | 65.24 | 71.91 | 66.38 | 67.67 | 73.10 | 55.38 | 64.06 | 65.21 | 58.93 | 65.24 | 71.91 | 66.38 | 67.67 | 73.10 | 55.38 | 64.06 | 3DShot2 | 47 | ||||||||||||||||||
| 48 | HHpred2 | 64.59 | 57.30 | 71.18 | 71.66 | 65.39 | 69.26 | 83.23 | 76.90 | 81.32 | 0.02 | 0.13 | 0.04 | 0.36 | 64.59 | 57.30 | 71.18 | 71.66 | 65.39 | 69.26 | 83.23 | 76.90 | 81.32 | 0.13 | HHpred2 | 48 | |||||||||||||
| 49 | Phyre2 | 64.50 | 56.79 | 68.96 | 71.53 | 64.69 | 66.27 | 83.23 | 74.58 | 81.31 | 0.13 | 0.36 | 71.35 | 65.90 | 77.04 | 78.47 | 73.27 | 77.02 | 86.39 | 83.86 | 81.77 | 0.33 | 0.54 | 0.54 | 0.55 | 0.38 | 0.65 | 0.35 | 0.57 | 0.20 | Phyre2 | 49 | |||||||
| 50 | SAM−T06−server | 64.15 | 52.76 | 72.53 | 71.41 | 63.66 | 71.83 | 85.76 | 81.33 | 81.59 | 0.17 | 0.14 | 0.43 | 0.48 | 0.39 | 70.11 | 64.23 | 73.36 | 76.11 | 72.15 | 71.83 | 86.71 | 81.33 | 82.98 | 0.03 | 0.14 | 0.09 | 0.19 | 0.41 | 0.18 | 0.40 | SAM−T06−server | 50 | ||||||
| 51 | Pcons_local | 63.79 | 60.17 | 60.47 | 64.60 | 60.89 | 60.65 | 75.32 | 70.46 | 68.22 | 72.69 | 66.28 | 76.26 | 78.96 | 74.26 | 75.07 | 88.61 | 84.60 | 84.46 | 0.65 | 0.63 | 0.42 | 0.65 | 0.54 | 0.30 | 0.75 | 0.68 | 0.63 | Pcons_local | 51 | |||||||||
| 52 | SAM−T02−server | 63.43 | 57.68 | 58.99 | 63.49 | 59.28 | 58.85 | 75.32 | 73.00 | 67.65 | 71.08 | 64.62 | 75.12 | 77.23 | 73.27 | 73.77 | 87.66 | 84.70 | 83.67 | 0.26 | 0.23 | 0.23 | 0.31 | 0.38 | 0.07 | 0.58 | 0.70 | 0.51 | SAM−T02−server | 52 | |||||||||
| 53 | LOOPP_Server | 62.46 | 54.45 | 56.94 | 67.82 | 65.84 | 57.30 | 71.20 | 65.30 | 63.79 | 72.06 | 67.79 | 72.28 | 78.09 | 75.54 | 73.13 | 83.23 | 78.59 | 81.26 | 0.50 | 0.99 | 0.48 | 0.76 | 0.12 | LOOPP_Server | 53 | |||||||||||||
| 54 | Poing | 62.28 | 55.46 | 69.69 | 67.70 | 60.44 | 67.92 | 83.23 | 74.58 | 81.31 | 0.13 | 0.36 | 62.54 | 55.55 | 70.64 | 68.32 | 63.37 | 69.19 | 83.23 | 74.58 | 81.31 | 0.13 | Poing | 54 | |||||||||||||||
| 55 | Phragment | 61.39 | 54.03 | 69.90 | 66.83 | 59.49 | 68.52 | 83.23 | 74.58 | 81.31 | 0.13 | 0.36 | 64.41 | 58.84 | 71.57 | 70.92 | 64.56 | 70.44 | 83.23 | 74.58 | 81.31 | 0.13 | Phragment | 55 | |||||||||||||||
| 56 | Distill | 59.43 | 54.27 | 54.55 | 61.26 | 58.46 | 56.03 | 67.72 | 63.71 | 63.65 | 59.43 | 54.27 | 56.12 | 61.26 | 58.46 | 57.09 | 72.47 | 68.14 | 65.49 | Distill | 56 | ||||||||||||||||||
| 57 | FEIG | 59.25 | 56.29 | 73.33 | 77.47 | 69.97 | 75.23 | 79.75 | 61.92 | 74.41 | 0.25 | 0.59 | 0.23 | 0.53 | 65.57 | 59.58 | 73.33 | 77.47 | 69.97 | 75.30 | 79.75 | 75.21 | 76.29 | 0.35 | 0.34 | FEIG | 57 | ||||||||||||
| 58 | rehtnap | 54.80 | 48.99 | 45.70 | 57.43 | 51.40 | 45.21 | 62.98 | 60.87 | 55.42 | 54.80 | 48.99 | 45.70 | 57.43 | 51.40 | 45.21 | 62.98 | 60.87 | 55.42 | rehtnap | 58 | ||||||||||||||||||
| 59 | MUFOLD−Server | 50.36 | 45.14 | 43.14 | 54.45 | 48.52 | 45.39 | 51.90 | 38.82 | 51.53 | 50.45 | 45.34 | 43.16 | 54.58 | 48.76 | 45.41 | 51.90 | 38.82 | 51.53 | MUFOLD−Server | 59 | ||||||||||||||||||
| 60 | HHpred5 | 39.95 | 31.23 | 30.01 | 46.53 | 37.38 | 38.66 | 36.71 | 28.48 | 26.71 | 39.95 | 31.23 | 30.01 | 46.53 | 37.38 | 38.66 | 36.71 | 28.48 | 26.71 | HHpred5 | 60 | ||||||||||||||||||
| 61 | BioSerf | 38.17 | 28.59 | 60.52 | 47.15 | 39.15 | 60.74 | 72.47 | 67.83 | 69.43 | 38.17 | 28.59 | 60.52 | 47.15 | 39.15 | 60.74 | 72.47 | 67.83 | 69.43 | BioSerf | 61 | ||||||||||||||||||
| 62 | FUGUE_KM | 34.16 | 24.73 | 18.89 | 34.03 | 27.80 | 21.71 | 47.78 | 37.66 | 31.47 | 64.06 | 60.26 | 60.52 | 69.31 | 62.62 | 60.05 | 76.27 | 73.52 | 69.93 | FUGUE_KM | 62 | ||||||||||||||||||
| 63 | OLGAFS | 32.92 | 23.84 | 16.68 | 33.42 | 26.57 | 20.11 | 45.89 | 36.81 | 27.84 | 70.46 | 63.58 | 74.21 | 76.61 | 71.91 | 72.82 | 86.71 | 77.64 | 82.98 | 0.12 | 0.09 | 0.18 | 0.15 | 0.41 | 0.40 | OLGAFS | 63 | ||||||||||||
| 64 | FOLDpro | 22.24 | 17.97 | 22.65 | 29.95 | 24.09 | 22.97 | 44.94 | 35.02 | 42.08 | 33.27 | 26.34 | 31.96 | 35.89 | 32.76 | 35.77 | 48.73 | 40.30 | 44.98 | FOLDpro | 64 | ||||||||||||||||||
| 65 | mariner1 | 15.39 | 10.85 | 11.06 | 19.55 | 14.52 | 20.47 | 17.72 | 12.87 | 7.30 | 57.03 | 52.64 | 53.22 | 63.49 | 58.13 | 60.75 | 49.37 | 42.41 | 40.99 | mariner1 | 65 | ||||||||||||||||||
| 66 | RBO−Proteus | 12.01 | 8.45 | 28.50 | 16.58 | 11.63 | 30.35 | 37.34 | 24.68 | 45.90 | 13.26 | 11.48 | 28.50 | 16.58 | 12.95 | 30.35 | 43.04 | 34.07 | 45.90 | RBO−Proteus | 66 | ||||||||||||||||||
| 67 | MUFOLD−MD | 11.30 | 9.85 | 28.73 | 15.72 | 14.03 | 32.96 | 34.49 | 27.11 | 39.59 | 14.50 | 11.80 | 31.36 | 20.17 | 15.14 | 34.06 | 34.49 | 31.22 | 48.78 | MUFOLD−MD | 67 | ||||||||||||||||||
| 68 | RANDOM | 9.49 | 8.26 | 9.49 | 12.48 | 10.50 | 12.48 | 28.91 | 24.80 | 28.91 | 9.49 | 8.26 | 9.49 | 12.48 | 10.50 | 12.48 | 28.91 | 24.80 | 28.91 | RANDOM | 68 | ||||||||||||||||||
| 69 | schenk−torda−server | 8.19 | 6.82 | 8.52 | 11.51 | 9.78 | 13.66 | 22.47 | 19.09 | 19.15 | 8.19 | 6.82 | 8.52 | 11.51 | 9.78 | 13.66 | 22.47 | 20.46 | 19.39 | schenk−torda−server | 69 | ||||||||||||||||||
| 70 | BHAGEERATH | BHAGEERATH | 70 | ||||||||||||||||||||||||||||||||||||
| 71 | Frankenstein | Frankenstein | 71 | ||||||||||||||||||||||||||||||||||||
| 72 | huber−torda−server | huber−torda−server | 72 | ||||||||||||||||||||||||||||||||||||
| 73 | mahmood−torda−server | mahmood−torda−server | 73 | ||||||||||||||||||||||||||||||||||||
| 74 | test_http_server_01 | test_http_server_01 | 74 | ||||||||||||||||||||||||||||||||||||