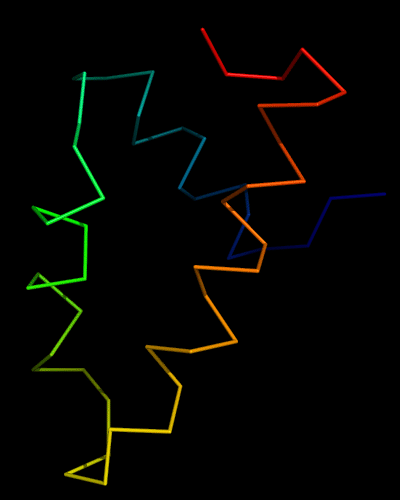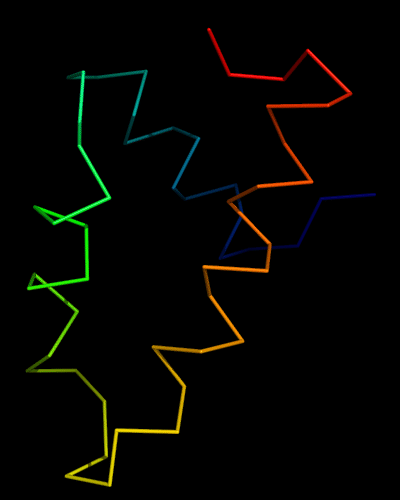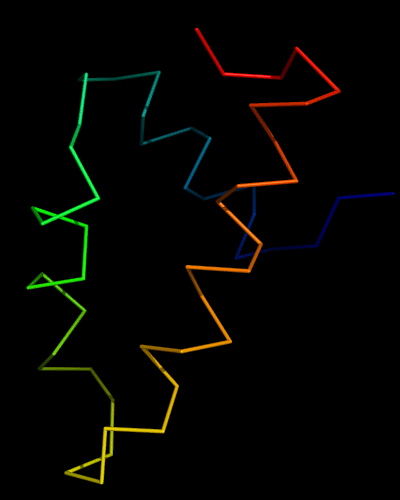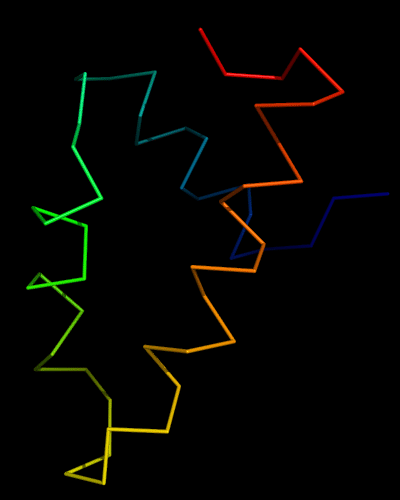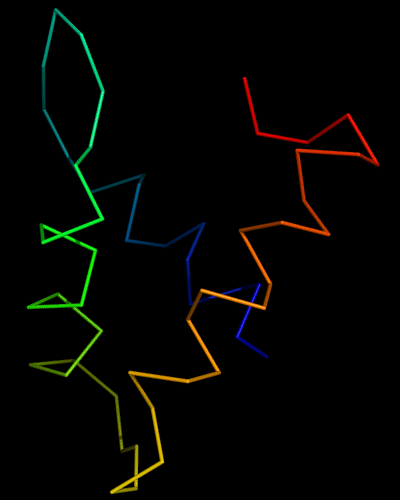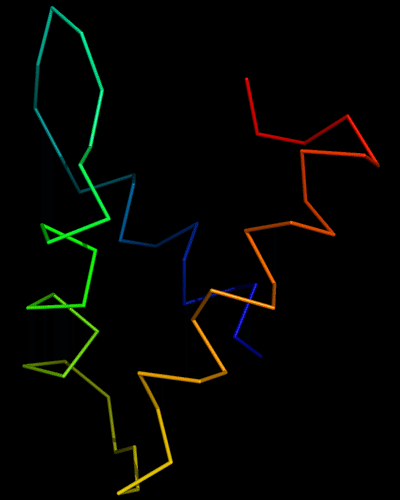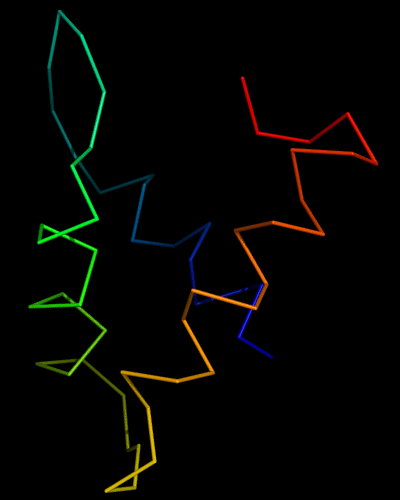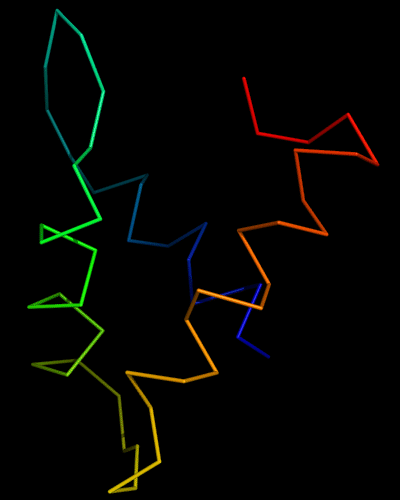T0443
putative regulatory protein (YP_208969.1) from Neisseria gonorrhoeae
Target sequence:
>T0443 FP12834A, Neisseria gonorrhoeae, 248 residues
MQPETSAQYQHRFSQAIRGGEAADGLPQDRLNVYIRLIRNNIHSFIDRCYTETRQYFDSKEWSRLKEGFVRDARAQTPYFQEIPGEFLQYCQSPPLSDGILALMDFEYTQLLAEVAQIPDIPDIHYSNDSKYTPSPAAFIRQYRYDVTHDLQEAETALLIWRNAEDDVMYQTLDGFDMMLLEIMGSSALSFDTLAQTLVEFMPKADNWKNILLGKWSGWIEQRIIIPSLSAISENMEGNSPSQNHLSA
Structure:
Determined by:
JCSG
PDB ID: 3dee
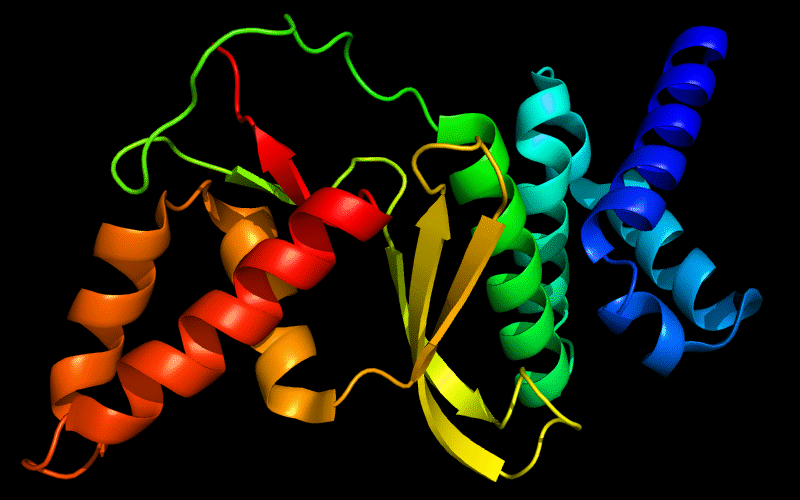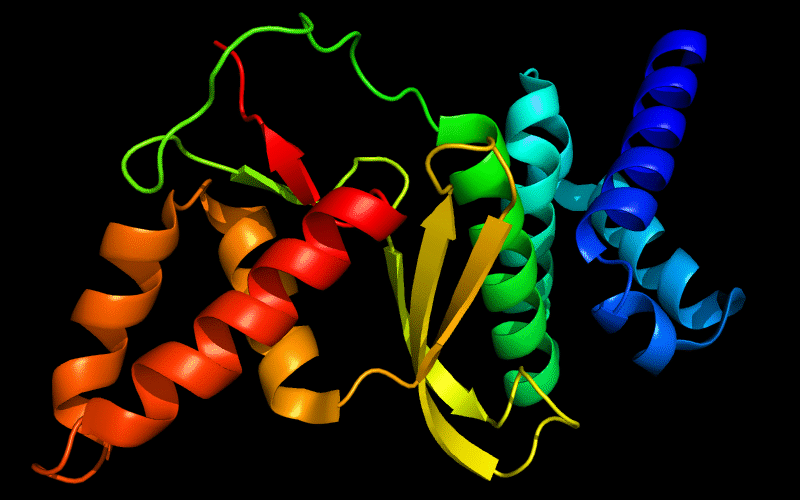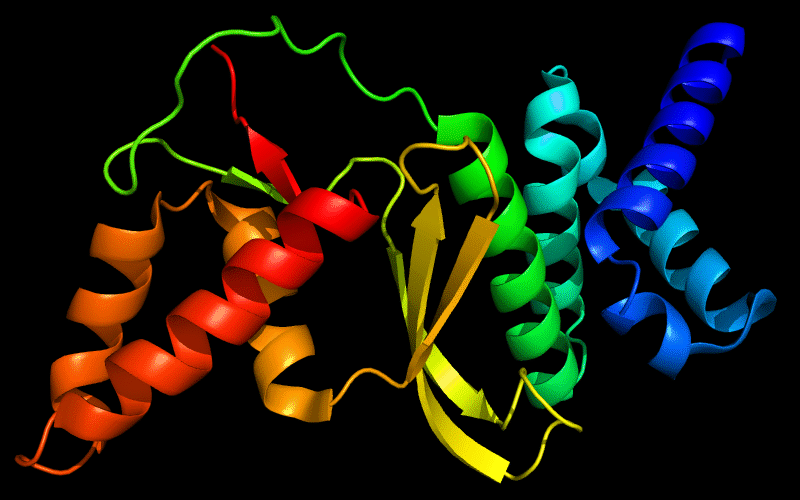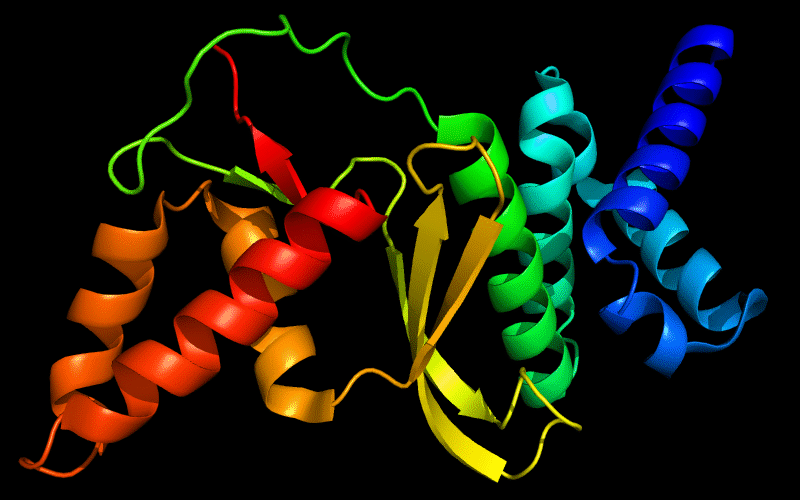
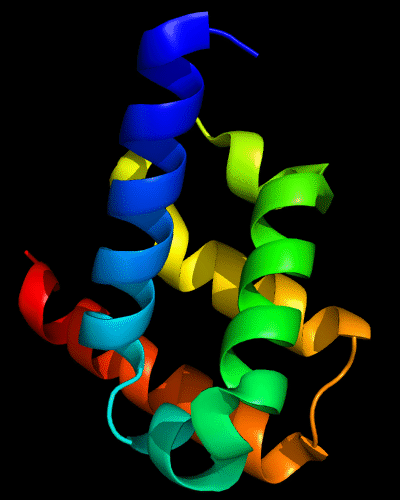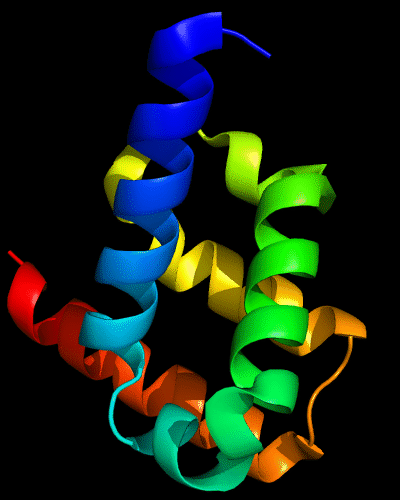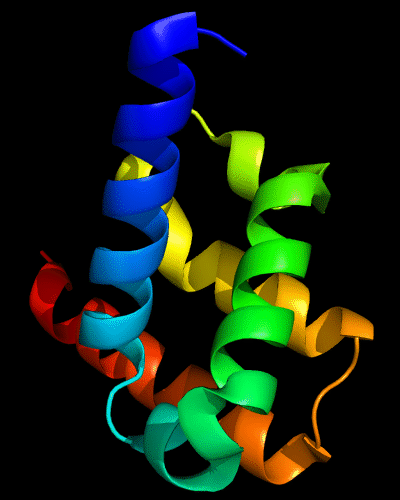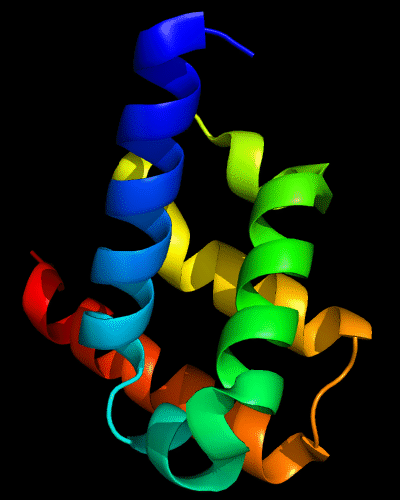
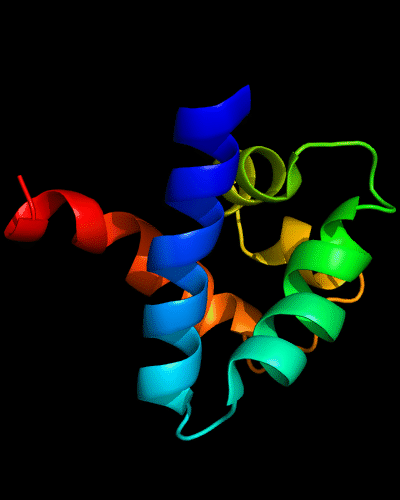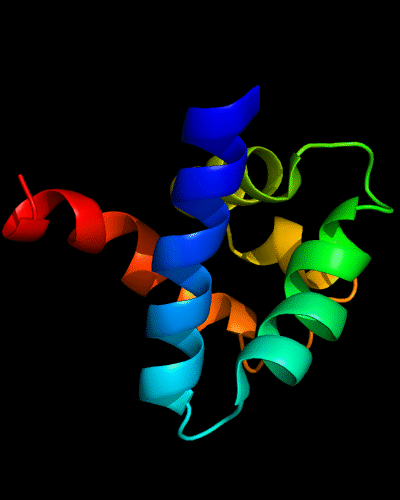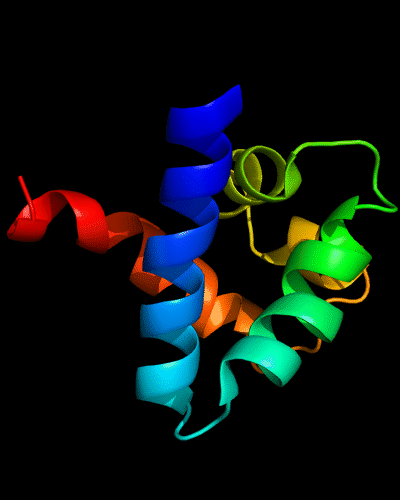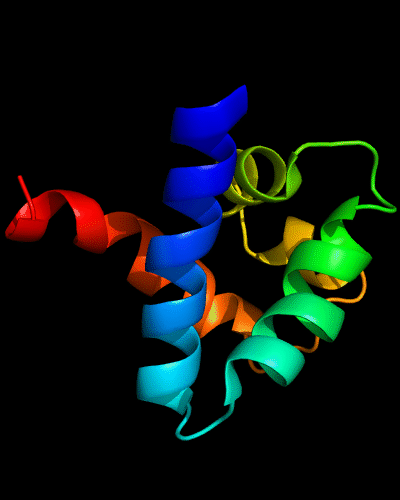
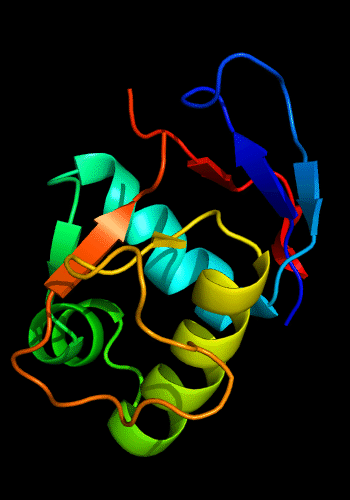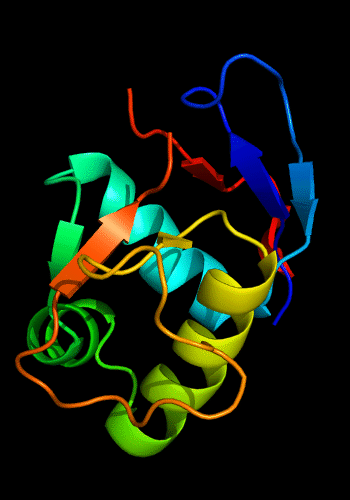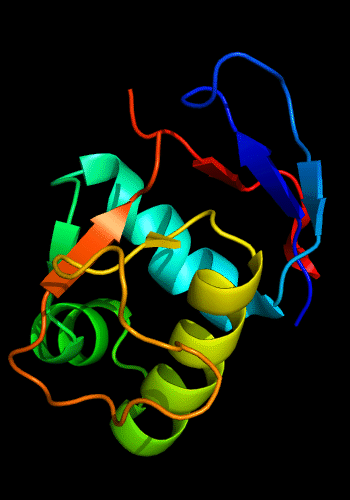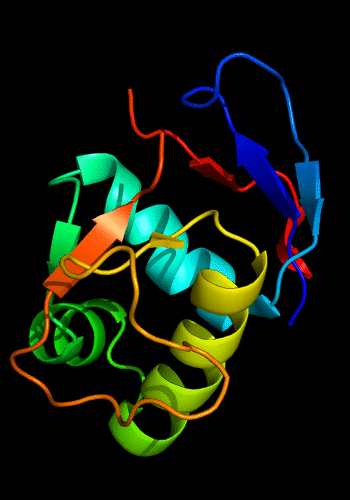
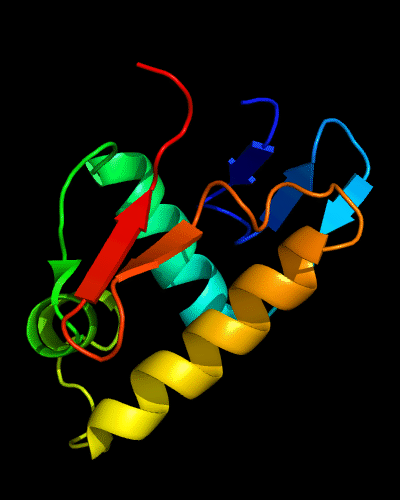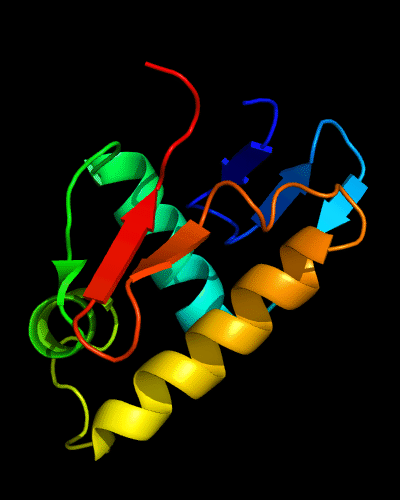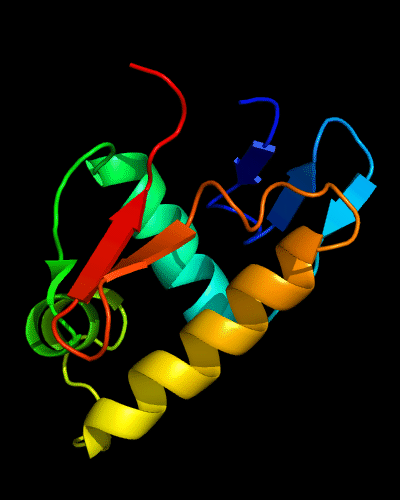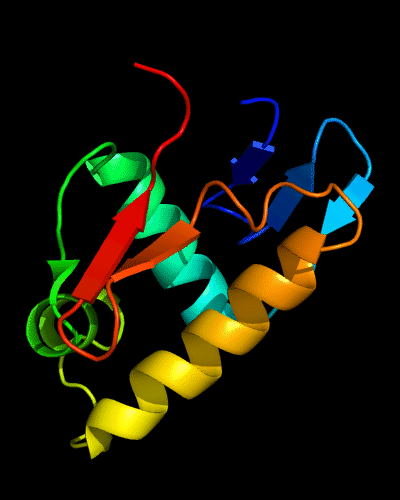
Cartoon diagram of 443: 3dee
Domains: PyMOL of domains
Two domains. Residue ranges in PDB: 31-116 and 117-230. Residue ranges in target: 31-116 and 117-230:
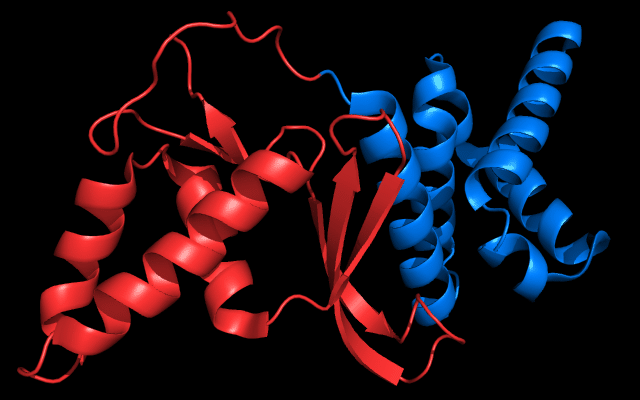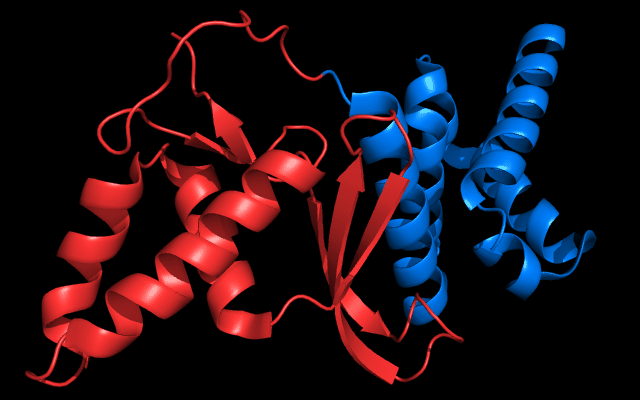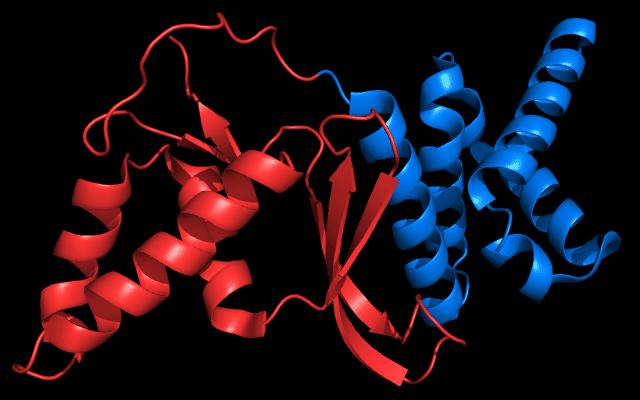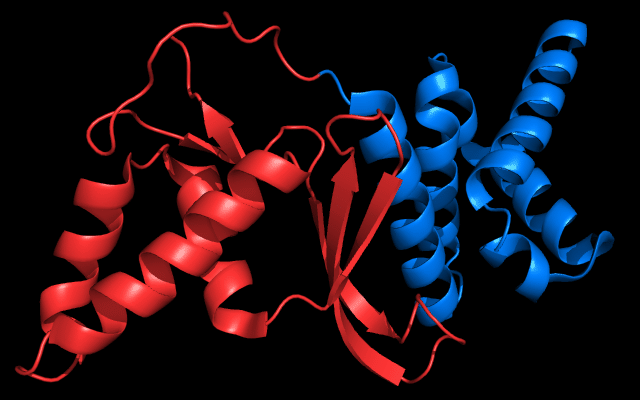
Cartoon diagram of 443 evolutionary domains: 3dee,
N- and C-domains are colored blue and red.
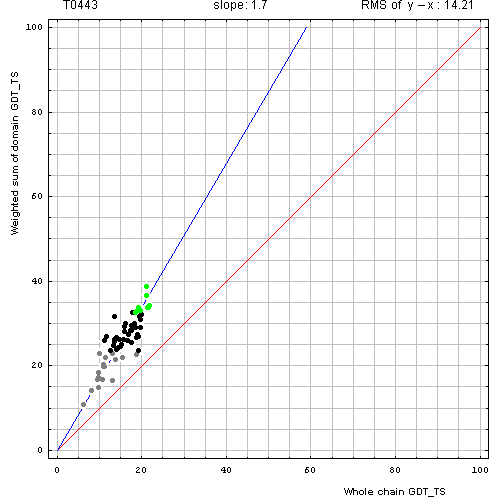
Correlation between weighted by the number of residues sum of GDT-TS scores for domain-based evlatuation (y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
To compute the weighted sum, GDT-TS for each domain was multiplied by the domain length, and this sum was divided by the sum of domain lengths. Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the weighted sum of domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
N-domain is from the SAM-domain (HhH motif) fold, both HhH motifs deteriorated, but the loops typically housing them are still possibly functional for binding.
Cartoon diagram of N-domain of T0443: 3dee residues 31-117
Middle domain of eIFα: 2aho chain B residues 96-176
DALI finds partial, but quite significant (Z-score >5) similarity to the cyclin fold, matching 4 out of 5 cyclin helices. If found, this template would produce a very accurate starting model:

Cartoon diagram of N-domain of T0443: 3dee residues 31-117
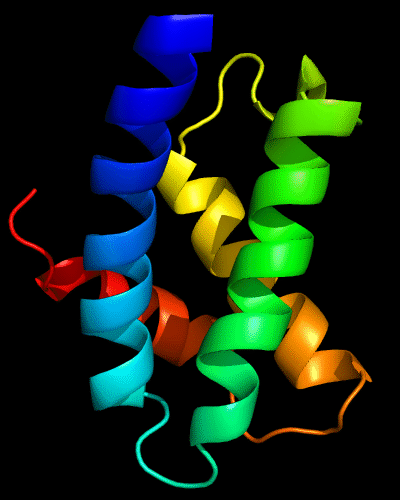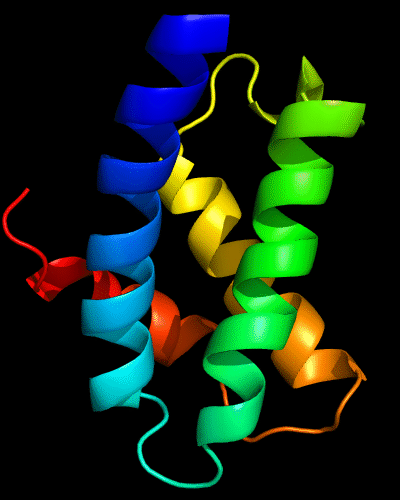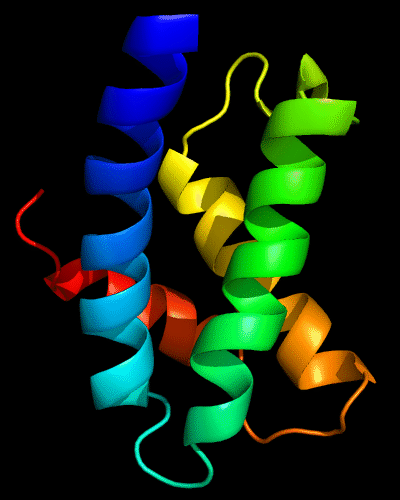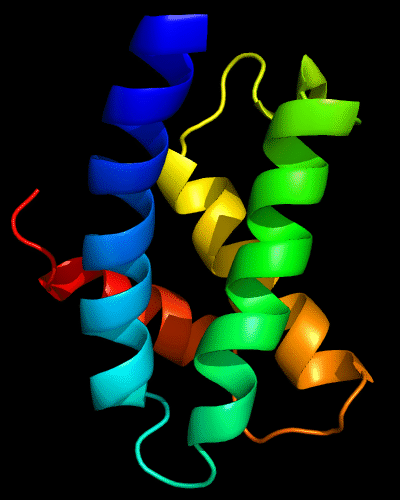
4 helices from a cyclin domain: 1gh6 chain B 648-733
The C-domain is a circularly permuted winged HTH, i.e. the last strand of the "wing" is the first strand of the domain, like in Methionine aminopeptidase:
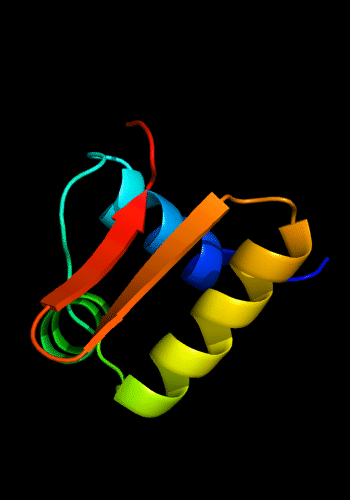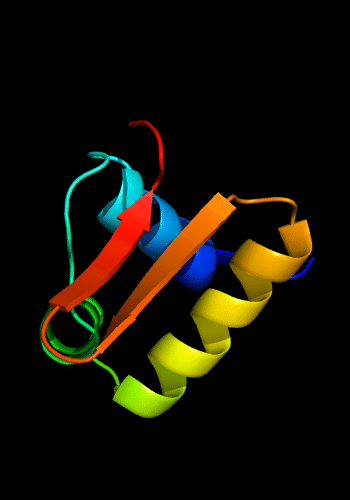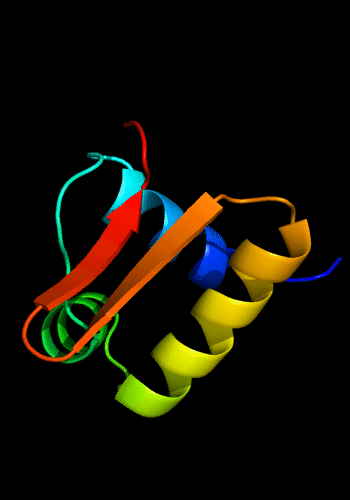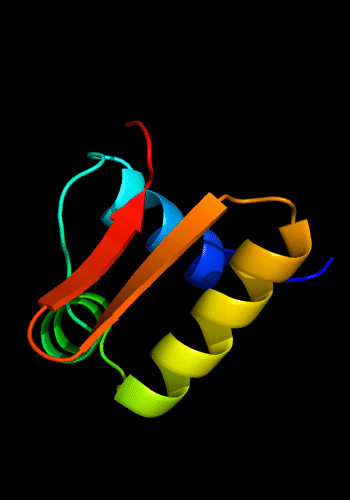
Classic winged HTH in biotin repressor: 1bia residues 1-63,
HTH helices are green and yellow, "wing" strands
are orange and red
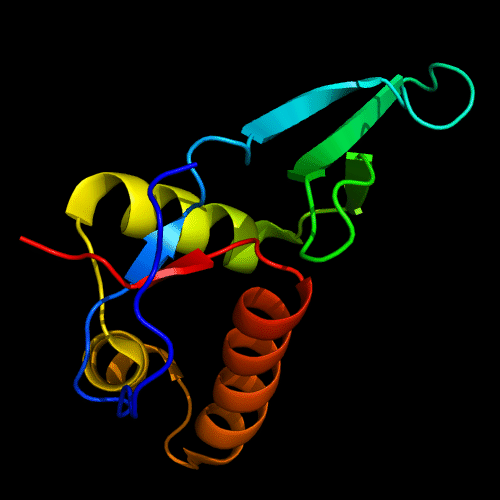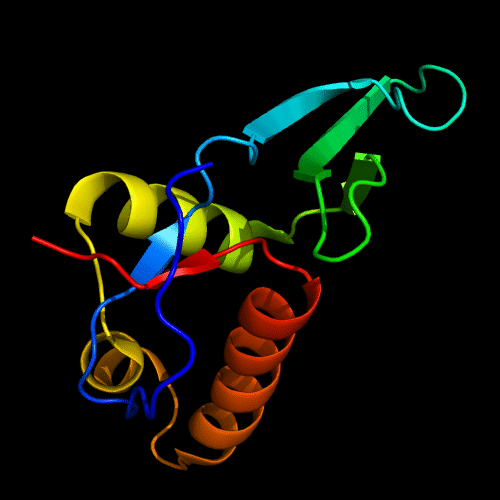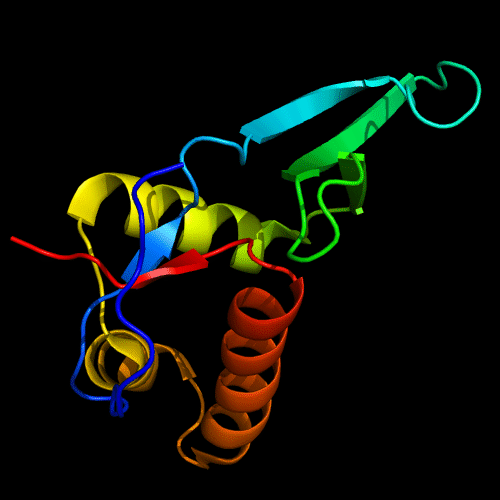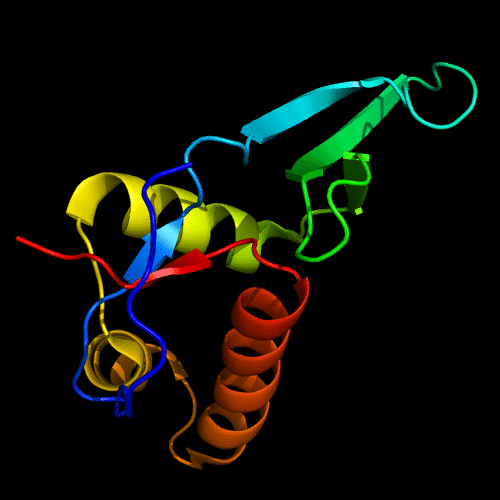
Cartoon diagram of C-domain of T0443: 3dee residues 118-230,
HTH helices are orange-yellow and orange, "wing" strands are blue and red
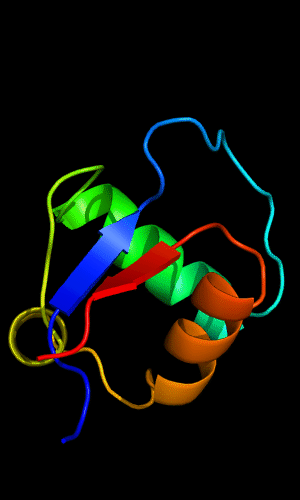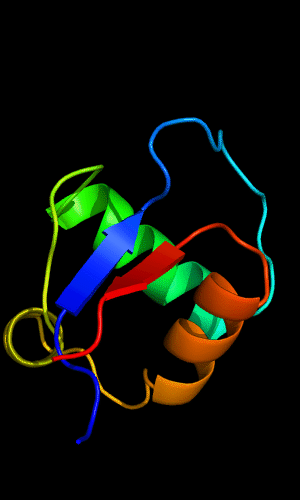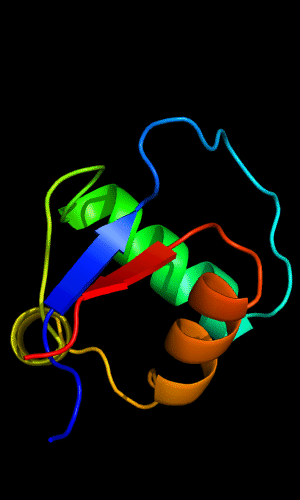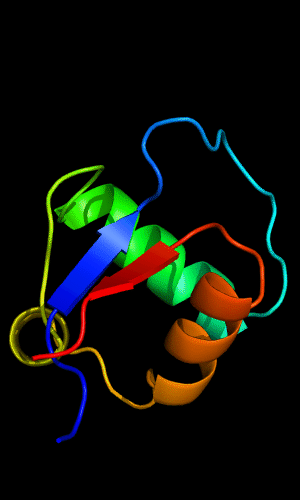
Circularly permuted HTH in Met aminopeptidase: 1b6a 378-446,
HTH helices are yellow and orange, "wing" strands are blue and red
Additionally, the winged HTH in the C-domain is decorated with a 3-stranded sheet inserted after the "wing". β-hairpins and strands are known to be present just before the N-helix of the HHTH domains, e.g. cullin 1st HTH and PhoB-like domains:

Cartoon diagram of C-domain of T0443: 3dee residues 118-230,
HTH helices are orange-yellow and orange, "wing" strands are blue and red,
side β-sheet is cyan-green
2nd HTH in cullin: 1ldj chains A:586-673, B:19-28, HTH helices are green and lime,
"wing" strands are yellow and orange,
side β-sheet is red and blue

Cartoon diagram of C-domain of T0443: 3dee residues 118-230,
HTH helices are orange-yellow and orange, "wing" strands are blue and red,
side β-sheet is cyan-green
HTH domain of PhoB: 1qqi residues 10-104
HTH helices are green and yellow-orange, "wing" strands are orange and red,
side β-sheet is blue-cyan
The HTH conformation looks a bit unusual, probably due to strong crystal contacts that bend the α-helices. The molecule is probably a dimer, it binds a small ligand with the two consreved Arg (one is not even modeled in the structure, it is just N-terminal to the first modeled residue), each from each monomer. This protein family is very large, and this kind of HTH without the N-domain exists as a separate protein: "Coenzyme PQQ synthesis protein D".
Interestingly HHpred server finds HTH domains as templates for this C-domain.
CASP category:
Whole chain: Free modeling.
1st domain: Fold recognition. 2nd domain: Fold recognition.
Closest templates:
The first COMPASS hit in SCOP database is to the DNA/RNA-binding 3-helical bundle HTH-containing superfamily:
Subject = d1hsta_ a.4.5.13 Histone H5, globular domain length=74 filtered_length=74 Neff=12.554 Smith-Waterman score = 56 Evalue = 3.65e-01 QUERY 172 TLDGFDMMLLEIMGS=SALSFDTLAQTLVEFMPKADNWKNILLGKWSGWIEQRIII 226 CONSENSUS_1 172 PLSPLEAALLQALAE=GGLSLEALLEALAEELPDFDALLALLAQLLAQLLDAGLIL 226 P+S++ + +++AL E +G+SL A+ +++ ++D++ +++LL++ L++L+++G+++ CONSENSUS_2 4 PYSEMILEAIKALKERKGSSLAAIKKYIEAKYKDVENFKSLLKRALKKLVAKGKLV 59 d1hsta_ 4 TYSEMIAAAIRAEKSRGGSSRQSIQKYIKSHYKVGHNADLQIKLSIRRLLAAGVLK 59
Although E-value estimate is only marginally significant (~0.4), the alignment is largely correct and can be used for template-based modeling of the C-domain. Here is structure comparison of the segments aligned by COMPASS, HTH motif is from green to red :
2dql was found by HHpred for HTH C-domain.
Target sequence - PDB file inconsistencies:
T0443 3dee.pdb T0443.pdb PyMOL PyMOL of domains
T0443 1 MQPETSAQYQHRFSQAIRGGEAADGLPQDRLNVYIRLIRNNIHSFIDRCYTETRQYFDSKEWSRLKEGFVRDARAQTPYFQEIPGEFLQYCQSPPLSDGILALMDFEYTQLLAEVAQIPDIPDIHYSNDSKYTPSPAAFIRQYRYDVTHDLQEAETALLIWRNAEDDVMYQTLDGFDMMLLEIMGSSALSFDTLAQTLVEFMPKADNWKNILLGKWSGWIEQRIIIPSLSAISENMEGNSPSQNHLSA 248 ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||:||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~~~~~~~~~ 3deeA 31 ------------------------------LNVYIRLIRNNIHSFIDRCYTETRQYFDSKEWSRLKEGFVRDARAQTPYFQEIPGEFLQYCQSLPLSDGILALMDFEYTQLLAEVAQIPDIPDIHYSNDSKYTPSPAAFIRQYRYDVTHDLQEAETALLIWRNAEDDVMYQTLDGFDMMLLEIMGSSALSFDTLAQTLVEFMPKADNWKNILLGKWSGWIEQRIIIPSLS------------------ 230
Residue change log: change 94 L to P; change 104, 169, 178, 179, 184, 202, MSE to MET;
2-domain protein, both domains used in evaluation:
1st domain: target 31-116 ; pdb 31-116
2nd domain: target 117-230 ; pdb 117-230
Sequence classification:
COG3219 in CDD.
Server predictions:
T0443:pdb 31-230:seq 31-230:FM; alignment
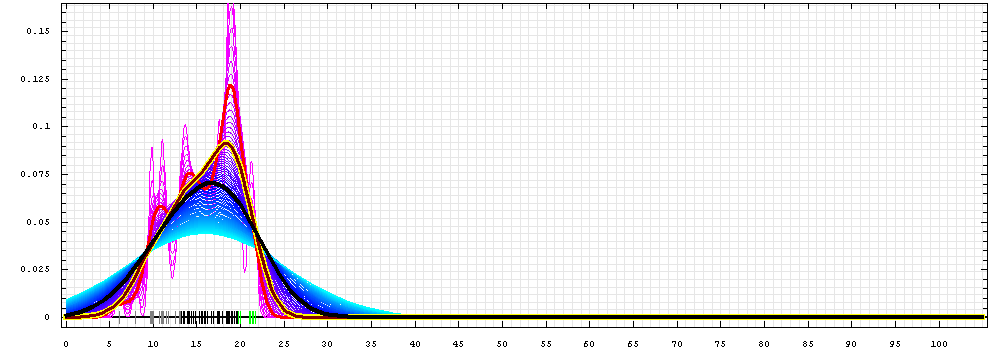
First models for T0443:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
443_1:pdb 31-116:seq 31-116:FR; alignment
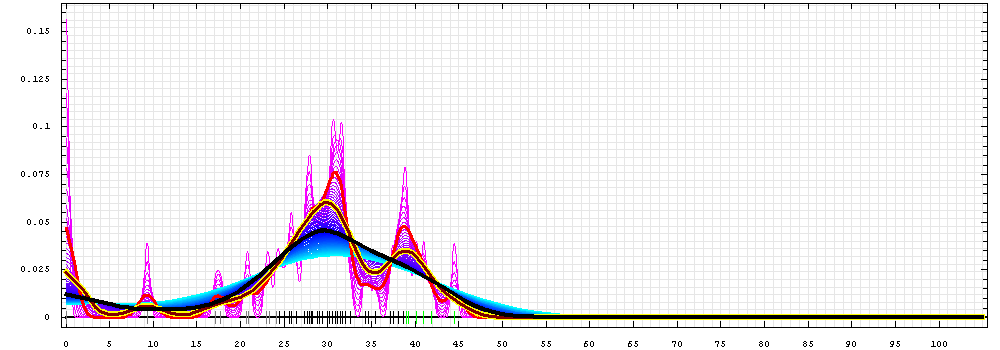
First models for T0443_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
443_2:pdb 117-230:seq 117-230:FR; alignment
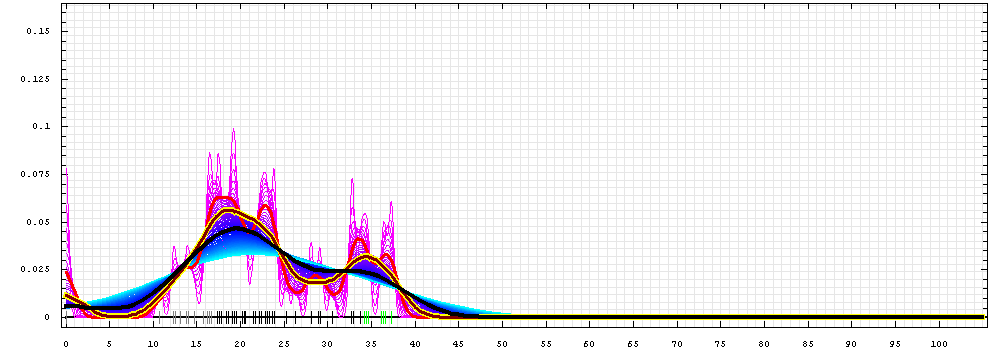
First models for T0443_2:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0443 | 443_1 | 443_2 | T0443 | 443_1 | 443_2 | T0443 | 443_1 | 443_2 | T0443 | 443_1 | 443_2 | ||||||||||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||||||||||||||
| 1 | fais@hgc | 23.50 | 21.08 | 35.95 | 34.59 | 28.20 | 46.88 | 39.47 | 34.80 | 38.69 | 2.20 | 2.72 | 0.89 | 0.40 | 0.09 | 1.08 | 2.05 | 2.48 | 0.69 | 23.50 | 21.12 | 36.22 | 39.24 | 31.88 | 46.88 | 39.47 | 34.94 | 39.99 | 1.70 | 2.04 | 0.74 | 0.78 | 0.35 | 0.95 | 1.47 | 1.79 | 0.50 | fais@hgc | 1 |
| 2 | 3DShot1 | 22.38 | 15.38 | 19.31 | 31.11 | 24.52 | 5.32 | 39.47 | 27.19 | 32.94 | 1.82 | 0.42 | 2.05 | 1.10 | 0.07 | 22.38 | 15.38 | 19.31 | 31.11 | 24.52 | 5.32 | 39.47 | 27.19 | 32.94 | 1.33 | 1.47 | 0.48 | 3DShot1 | 2 | ||||||||||
| 3 | LevittGroup | 21.75 | 16.58 | 38.64 | 35.47 | 32.66 | 45.90 | 38.38 | 28.87 | 43.36 | 1.61 | 0.90 | 1.13 | 0.55 | 0.97 | 1.02 | 1.89 | 1.40 | 1.20 | 24.62 | 19.25 | 40.35 | 38.37 | 35.66 | 47.12 | 42.33 | 32.24 | 45.48 | 2.07 | 1.32 | 1.17 | 0.62 | 1.16 | 0.96 | 1.87 | 1.33 | 1.15 | LevittGroup | 3 |
| 4 | SAM−T08−server | 21.75 | 16.42 | 37.00 | 30.81 | 27.91 | 36.53 | 36.62 | 27.56 | 45.37 | 1.61 | 0.84 | 0.98 | 0.03 | 0.41 | 1.64 | 1.17 | 1.42 | 21.75 | 17.29 | 37.50 | 32.56 | 28.59 | 41.14 | 37.06 | 30.34 | 45.37 | 1.12 | 0.57 | 0.88 | 0.51 | 1.13 | 1.01 | 1.14 | SAM−T08−server | 4 | |||
| 5 | FAMSD | 21.50 | 16.54 | 27.65 | 29.07 | 29.07 | 30.18 | 37.28 | 30.04 | 33.39 | 1.53 | 0.89 | 0.14 | 0.26 | 1.73 | 1.62 | 0.11 | 21.50 | 16.54 | 27.65 | 35.17 | 29.07 | 33.74 | 37.28 | 30.04 | 33.39 | 1.04 | 0.28 | 0.03 | 1.16 | 0.96 | FAMSD | 5 | ||||||
| 6 | circle | 21.38 | 15.58 | 27.79 | 29.36 | 28.78 | 30.23 | 37.28 | 26.90 | 33.61 | 1.49 | 0.50 | 0.15 | 0.20 | 1.73 | 1.05 | 0.14 | 21.62 | 17.83 | 28.31 | 29.65 | 29.07 | 31.31 | 37.50 | 30.85 | 33.90 | 1.08 | 0.78 | 1.19 | 1.10 | circle | 6 | |||||||
| 7 | DBAKER | 21.12 | 18.88 | 39.49 | 29.36 | 26.16 | 43.73 | 37.28 | 31.87 | 45.75 | 1.40 | 1.83 | 1.21 | 0.88 | 1.73 | 1.95 | 1.47 | 21.12 | 18.88 | 40.19 | 36.34 | 32.27 | 47.28 | 37.28 | 31.87 | 45.75 | 0.92 | 1.18 | 1.15 | 0.25 | 0.44 | 0.98 | 1.16 | 1.27 | 1.18 | DBAKER | 7 | ||
| 8 | BAKER−ROBETTA | 21.00 | 19.33 | 42.13 | 41.86 | 37.40 | 45.93 | 36.40 | 31.73 | 48.88 | 1.36 | 2.02 | 1.44 | 1.65 | 1.90 | 1.02 | 1.61 | 1.92 | 1.81 | 22.88 | 21.46 | 42.13 | 41.86 | 37.40 | 52.02 | 39.47 | 36.99 | 48.88 | 1.50 | 2.17 | 1.35 | 1.26 | 1.54 | 1.34 | 1.47 | 2.14 | 1.55 | BAKER−ROBETTA | 8 |
| 9 | Zico | 21.00 | 19.25 | 42.10 | 41.86 | 37.21 | 46.32 | 36.18 | 30.56 | 48.65 | 1.36 | 1.98 | 1.44 | 1.65 | 1.86 | 1.05 | 1.58 | 1.71 | 1.78 | 21.00 | 19.25 | 42.10 | 41.86 | 37.21 | 46.91 | 36.18 | 30.56 | 48.65 | 0.88 | 1.32 | 1.35 | 1.26 | 1.49 | 0.95 | 1.01 | 1.05 | 1.52 | Zico | 9 |
| 10 | ZicoFullSTP | 21.00 | 19.25 | 42.10 | 41.86 | 37.21 | 46.32 | 36.18 | 30.56 | 48.65 | 1.36 | 1.98 | 1.44 | 1.65 | 1.86 | 1.05 | 1.58 | 1.71 | 1.78 | 21.00 | 19.25 | 42.10 | 41.86 | 37.21 | 46.91 | 36.18 | 30.56 | 48.65 | 0.88 | 1.32 | 1.35 | 1.26 | 1.49 | 0.95 | 1.01 | 1.05 | 1.52 | ZicoFullSTP | 10 |
| 11 | SAM−T08−human | 21.00 | 17.00 | 41.42 | 41.86 | 36.63 | 46.16 | 37.06 | 31.07 | 47.60 | 1.36 | 1.07 | 1.38 | 1.65 | 1.75 | 1.04 | 1.70 | 1.80 | 1.67 | 21.25 | 19.88 | 43.62 | 42.15 | 37.40 | 49.24 | 37.06 | 31.36 | 49.52 | 0.96 | 1.56 | 1.50 | 1.31 | 1.54 | 1.13 | 1.13 | 1.19 | 1.63 | SAM−T08−human | 11 |
| 12 | PSI | 21.00 | 16.00 | 38.97 | 37.21 | 34.50 | 48.33 | 36.18 | 29.75 | 42.64 | 1.36 | 0.67 | 1.16 | 0.85 | 1.33 | 1.18 | 1.58 | 1.56 | 1.13 | 24.62 | 19.58 | 39.77 | 37.79 | 34.50 | 48.78 | 41.67 | 32.16 | 44.69 | 2.07 | 1.45 | 1.11 | 0.51 | 0.91 | 1.09 | 1.78 | 1.32 | 1.06 | PSI | 12 |
| 13 | fams−ace2 | 20.88 | 15.88 | 38.09 | 36.34 | 33.62 | 45.89 | 36.40 | 27.05 | 42.45 | 1.32 | 0.62 | 1.08 | 0.70 | 1.16 | 1.02 | 1.61 | 1.07 | 1.11 | 20.88 | 17.04 | 38.09 | 40.70 | 37.02 | 45.89 | 36.40 | 27.05 | 42.45 | 0.84 | 0.47 | 0.94 | 1.05 | 1.45 | 0.87 | 1.04 | 0.45 | 0.79 | fams−ace2 | 13 |
| 14 | FAMS−multi | 20.75 | 18.83 | 27.52 | 28.78 | 28.78 | 30.45 | 36.18 | 32.82 | 33.16 | 1.28 | 1.81 | 0.13 | 0.20 | 0.01 | 1.58 | 2.12 | 0.09 | 21.25 | 19.83 | 27.52 | 37.79 | 31.30 | 30.45 | 36.84 | 34.06 | 33.77 | 0.96 | 1.54 | 0.51 | 0.23 | 1.10 | 1.64 | FAMS−multi | 14 | ||||
| 15 | IBT_LT | 20.62 | 18.21 | 16.25 | 40.99 | 32.46 | 17.16 | 35.97 | 31.73 | 22.22 | 1.24 | 1.56 | 1.50 | 0.93 | 1.55 | 1.92 | 20.62 | 18.21 | 16.25 | 40.99 | 32.46 | 17.16 | 35.97 | 31.73 | 22.22 | 0.75 | 0.92 | 1.10 | 0.48 | 0.98 | 1.25 | IBT_LT | 15 | ||||||
| 16 | ABIpro | 20.62 | 17.67 | 40.19 | 39.24 | 34.01 | 40.75 | 36.40 | 31.87 | 48.39 | 1.24 | 1.34 | 1.27 | 1.20 | 1.23 | 0.68 | 1.61 | 1.95 | 1.75 | 24.62 | 19.33 | 40.19 | 39.24 | 34.01 | 48.78 | 40.57 | 31.87 | 48.39 | 2.07 | 1.35 | 1.15 | 0.78 | 0.81 | 1.09 | 1.63 | 1.27 | 1.49 | ABIpro | 16 |
| 17 | Bilab−UT | 20.50 | 17.75 | 35.75 | 36.92 | 30.52 | 43.07 | 35.53 | 30.70 | 40.12 | 1.20 | 1.38 | 0.87 | 0.80 | 0.55 | 0.83 | 1.48 | 1.73 | 0.85 | 23.62 | 19.33 | 42.13 | 41.86 | 37.40 | 48.33 | 41.67 | 31.73 | 48.88 | 1.74 | 1.35 | 1.35 | 1.26 | 1.54 | 1.06 | 1.78 | 1.25 | 1.55 | Bilab−UT | 17 |
| 18 | Jiang_Zhu | 20.25 | 17.00 | 27.94 | 47.09 | 39.73 | 40.09 | 24.56 | 17.98 | 29.10 | 1.11 | 1.07 | 0.17 | 2.56 | 2.36 | 0.64 | 20.25 | 17.00 | 27.94 | 47.09 | 39.73 | 40.09 | 24.56 | 20.39 | 31.12 | 0.63 | 0.46 | 2.22 | 2.03 | 0.43 | Jiang_Zhu | 18 | |||||||
| 19 | FALCON_CONSENSUS | 20.00 | 15.46 | 39.54 | 44.48 | 35.17 | 43.00 | 22.81 | 19.74 | 45.67 | 1.03 | 0.45 | 1.21 | 2.10 | 1.46 | 0.83 | 1.46 | 20.00 | 15.46 | 39.54 | 44.48 | 35.17 | 45.59 | 24.56 | 21.13 | 45.67 | 0.55 | 1.09 | 1.74 | 1.06 | 0.85 | 1.17 | FALCON_CONSENSUS | 19 | |||||
| 20 | FALCON | 20.00 | 15.46 | 39.54 | 44.48 | 35.17 | 43.00 | 22.81 | 19.74 | 45.67 | 1.03 | 0.45 | 1.21 | 2.10 | 1.46 | 0.83 | 1.46 | 20.50 | 17.75 | 39.54 | 44.48 | 35.17 | 45.59 | 35.53 | 30.70 | 46.12 | 0.71 | 0.75 | 1.09 | 1.74 | 1.06 | 0.85 | 0.92 | 1.07 | 1.23 | FALCON | 20 | ||
| 21 | SAINT1 | 19.88 | 16.83 | 38.66 | 46.22 | 40.50 | 50.50 | 28.07 | 20.61 | 40.93 | 0.99 | 1.01 | 1.13 | 2.41 | 2.51 | 1.32 | 0.42 | 0.94 | 19.88 | 16.83 | 38.66 | 46.22 | 40.50 | 50.50 | 28.07 | 20.61 | 40.93 | 0.51 | 0.39 | 0.99 | 2.06 | 2.20 | 1.22 | 0.61 | SAINT1 | 21 | |||
| 22 | METATASSER | 19.75 | 17.42 | 37.51 | 40.12 | 35.47 | 44.85 | 23.90 | 21.13 | 40.23 | 0.95 | 1.24 | 1.03 | 1.35 | 1.52 | 0.95 | 0.00 | 0.86 | 19.75 | 17.42 | 37.51 | 40.12 | 35.47 | 44.85 | 23.90 | 21.13 | 40.42 | 0.46 | 0.62 | 0.88 | 0.94 | 1.12 | 0.79 | 0.55 | METATASSER | 22 | |||
| 23 | MULTICOM−CLUSTER | 19.62 | 17.46 | 27.06 | 31.39 | 27.13 | 20.89 | 34.43 | 30.63 | 37.08 | 0.90 | 1.26 | 0.09 | 1.33 | 1.72 | 0.52 | 19.62 | 17.46 | 27.06 | 33.14 | 28.49 | 24.25 | 34.43 | 30.63 | 37.08 | 0.42 | 0.64 | 0.76 | 1.06 | 0.16 | MULTICOM−CLUSTER | 23 | |||||||
| 24 | pro−sp3−TASSER | 19.62 | 14.21 | 36.66 | 38.08 | 25.10 | 44.03 | 22.37 | 21.56 | 40.01 | 0.90 | 0.95 | 1.00 | 0.90 | 0.08 | 0.84 | 19.62 | 16.79 | 38.90 | 38.95 | 34.69 | 51.31 | 32.24 | 27.12 | 40.90 | 0.42 | 0.38 | 1.02 | 0.73 | 0.96 | 1.29 | 0.46 | 0.47 | 0.61 | pro−sp3−TASSER | 24 | |||
| 25 | TASSER | 19.50 | 16.21 | 37.12 | 36.63 | 31.98 | 46.25 | 22.59 | 21.27 | 39.83 | 0.86 | 0.75 | 0.99 | 0.75 | 0.83 | 1.04 | 0.03 | 0.82 | 21.12 | 19.54 | 40.92 | 43.02 | 40.50 | 50.72 | 36.84 | 24.85 | 44.71 | 0.92 | 1.43 | 1.23 | 1.47 | 2.20 | 1.24 | 1.10 | 0.08 | 1.06 | TASSER | 25 | |
| 26 | Phyre2 | 19.50 | 15.25 | 26.96 | 27.62 | 22.19 | 12.20 | 34.65 | 24.85 | 41.66 | 0.86 | 0.37 | 0.08 | 1.36 | 0.67 | 1.02 | 19.50 | 16.71 | 26.96 | 28.49 | 26.26 | 24.36 | 34.65 | 27.19 | 41.66 | 0.38 | 0.35 | 0.79 | 0.48 | 0.70 | Phyre2 | 26 | |||||||
| 27 | GS−MetaServer2 | 19.25 | 17.42 | 0.00 | 0.00 | 33.77 | 30.56 | 21.72 | 0.78 | 1.24 | 1.23 | 1.71 | 19.62 | 17.42 | 18.74 | 34.01 | 27.81 | 18.22 | 34.43 | 30.56 | 25.29 | 0.42 | 0.62 | 0.76 | 1.05 | GS−MetaServer2 | 27 | ||||||||||||
| 28 | GeneSilicoMetaServer | 19.25 | 17.42 | 0.00 | 0.00 | 33.77 | 30.56 | 21.72 | 0.78 | 1.24 | 1.23 | 1.71 | 19.62 | 17.42 | 18.74 | 34.01 | 27.81 | 18.22 | 34.43 | 30.56 | 25.29 | 0.42 | 0.62 | 0.76 | 1.05 | GeneSilicoMetaServer | 28 | ||||||||||||
| 29 | HHpred4 | 19.25 | 15.50 | 4.52 | 9.30 | 8.14 | 34.21 | 27.78 | 35.28 | 0.78 | 0.47 | 1.29 | 1.21 | 0.32 | 19.25 | 15.50 | 4.52 | 9.30 | 8.14 | 34.21 | 27.78 | 35.28 | 0.30 | 0.73 | 0.58 | HHpred4 | 29 | ||||||||||||
| 30 | Zhang | 19.25 | 14.92 | 40.54 | 36.34 | 31.88 | 49.78 | 32.90 | 25.15 | 44.38 | 0.78 | 0.23 | 1.30 | 0.70 | 0.81 | 1.27 | 1.11 | 0.73 | 1.32 | 21.50 | 20.00 | 40.80 | 44.19 | 36.53 | 53.36 | 37.28 | 34.65 | 45.63 | 1.04 | 1.61 | 1.21 | 1.69 | 1.35 | 1.44 | 1.16 | 1.74 | 1.17 | Zhang | 30 |
| 31 | Chicken_George | 19.25 | 14.58 | 35.03 | 34.88 | 30.23 | 42.86 | 33.55 | 25.36 | 38.83 | 0.78 | 0.10 | 0.80 | 0.45 | 0.49 | 0.82 | 1.20 | 0.77 | 0.71 | 19.25 | 15.04 | 38.56 | 37.79 | 34.98 | 49.15 | 33.55 | 25.36 | 44.41 | 0.30 | 0.98 | 0.51 | 1.02 | 1.12 | 0.64 | 0.17 | 1.02 | Chicken_George | 31 | |
| 32 | Bates_BMM | 19.12 | 17.62 | 36.00 | 38.95 | 34.30 | 43.33 | 30.92 | 26.39 | 40.05 | 0.74 | 1.32 | 0.89 | 1.15 | 1.29 | 0.85 | 0.82 | 0.95 | 0.84 | 24.12 | 18.75 | 39.29 | 38.95 | 34.30 | 43.58 | 42.76 | 35.82 | 46.28 | 1.91 | 1.13 | 1.06 | 0.73 | 0.87 | 0.69 | 1.93 | 1.94 | 1.24 | Bates_BMM | 32 |
| 33 | MULTICOM−RANK | 19.12 | 16.75 | 24.24 | 39.24 | 32.66 | 26.73 | 17.76 | 16.59 | 28.83 | 0.74 | 0.97 | 1.20 | 0.97 | 19.12 | 16.75 | 24.36 | 39.24 | 32.66 | 26.73 | 23.46 | 19.37 | 32.56 | 0.26 | 0.36 | 0.78 | 0.52 | MULTICOM−RANK | 33 | ||||||||||
| 34 | McGuffin | 19.00 | 17.29 | 35.26 | 36.63 | 32.85 | 49.72 | 25.44 | 15.50 | 34.47 | 0.70 | 1.19 | 0.82 | 0.75 | 1.00 | 1.27 | 0.04 | 0.23 | 21.75 | 17.29 | 37.00 | 36.63 | 32.85 | 49.72 | 36.62 | 30.56 | 45.37 | 1.12 | 0.57 | 0.82 | 0.30 | 0.56 | 1.16 | 1.07 | 1.05 | 1.14 | McGuffin | 34 | |
| 35 | MUFOLD−Server | 19.00 | 16.38 | 29.42 | 34.59 | 29.75 | 40.55 | 22.15 | 17.18 | 30.76 | 0.70 | 0.82 | 0.30 | 0.40 | 0.39 | 0.67 | 19.00 | 17.08 | 29.42 | 34.59 | 30.91 | 40.55 | 22.15 | 17.18 | 30.77 | 0.22 | 0.49 | 0.05 | 0.15 | 0.46 | MUFOLD−Server | 35 | |||||||
| 36 | Elofsson | 18.88 | 16.67 | 23.96 | 38.66 | 33.82 | 26.92 | 17.76 | 16.52 | 28.22 | 0.66 | 0.94 | 1.10 | 1.20 | 18.88 | 17.21 | 24.11 | 39.24 | 34.40 | 26.92 | 17.76 | 16.52 | 28.52 | 0.18 | 0.54 | 0.78 | 0.89 | Elofsson | 36 | ||||||||||
| 37 | MULTICOM−CMFR | 18.75 | 16.54 | 23.83 | 38.95 | 33.72 | 26.55 | 17.32 | 16.37 | 28.20 | 0.61 | 0.89 | 1.15 | 1.18 | 18.75 | 16.54 | 28.15 | 38.95 | 33.72 | 31.29 | 23.46 | 18.93 | 36.50 | 0.13 | 0.28 | 0.73 | 0.75 | 0.09 | MULTICOM−CMFR | 37 | |||||||||
| 38 | MULTICOM−REFINE | 18.75 | 16.50 | 23.99 | 38.66 | 33.43 | 26.46 | 17.54 | 16.16 | 28.53 | 0.61 | 0.87 | 1.10 | 1.12 | 19.38 | 17.38 | 28.28 | 38.66 | 33.82 | 27.59 | 34.65 | 31.43 | 37.11 | 0.34 | 0.60 | 0.67 | 0.77 | 0.79 | 1.20 | 0.16 | MULTICOM−REFINE | 38 | |||||||
| 39 | MUProt | 18.75 | 16.46 | 23.97 | 38.66 | 33.82 | 27.00 | 17.54 | 16.16 | 28.22 | 0.61 | 0.86 | 1.10 | 1.20 | 19.75 | 16.50 | 28.28 | 38.66 | 33.82 | 27.00 | 34.65 | 27.27 | 37.20 | 0.46 | 0.27 | 0.67 | 0.77 | 0.79 | 0.49 | 0.17 | MUProt | 39 | |||||||
| 40 | HHpred5 | 18.75 | 15.92 | 6.44 | 9.30 | 8.14 | 32.90 | 26.46 | 38.58 | 0.61 | 0.64 | 1.11 | 0.97 | 0.68 | 18.75 | 15.92 | 6.44 | 9.30 | 8.14 | 32.90 | 26.46 | 38.58 | 0.13 | 0.04 | 0.55 | 0.35 | 0.34 | HHpred5 | 40 | ||||||||||
| 41 | FFASsuboptimal | 18.75 | 13.62 | 0.00 | 0.00 | 32.90 | 23.90 | 24.08 | 0.61 | 1.11 | 0.50 | 19.25 | 16.75 | 0.00 | 0.00 | 33.77 | 29.39 | 26.48 | 0.30 | 0.36 | 0.67 | 0.85 | FFASsuboptimal | 41 | |||||||||||||||
| 42 | mufold | 18.62 | 17.62 | 32.12 | 36.34 | 33.62 | 42.46 | 24.34 | 17.91 | 33.17 | 0.57 | 1.32 | 0.54 | 0.70 | 1.16 | 0.79 | 0.09 | 21.00 | 19.33 | 42.30 | 41.86 | 37.40 | 46.05 | 36.18 | 30.56 | 49.07 | 0.88 | 1.35 | 1.37 | 1.26 | 1.54 | 0.88 | 1.01 | 1.05 | 1.57 | mufold | 42 | ||
| 43 | FFASstandard | 18.62 | 15.79 | 0.00 | 0.00 | 32.67 | 27.70 | 20.99 | 0.57 | 0.59 | 1.07 | 1.19 | 18.62 | 15.79 | 0.00 | 0.00 | 32.67 | 27.70 | 20.99 | 0.09 | 0.52 | 0.56 | FFASstandard | 43 | |||||||||||||||
| 44 | FFASflextemplate | 18.62 | 15.79 | 0.00 | 0.00 | 32.67 | 27.70 | 20.99 | 0.57 | 0.59 | 1.07 | 1.19 | 18.62 | 15.79 | 0.00 | 0.00 | 32.67 | 27.70 | 20.99 | 0.09 | 0.52 | 0.56 | FFASflextemplate | 44 | |||||||||||||||
| 45 | RBO−Proteus | 18.50 | 15.88 | 35.57 | 27.33 | 23.26 | 33.45 | 30.48 | 25.66 | 42.23 | 0.53 | 0.62 | 0.85 | 0.20 | 0.76 | 0.82 | 1.08 | 19.62 | 17.12 | 37.97 | 36.92 | 34.40 | 43.24 | 30.48 | 26.10 | 43.63 | 0.42 | 0.50 | 0.92 | 0.35 | 0.89 | 0.67 | 0.21 | 0.29 | 0.93 | RBO−Proteus | 45 | ||
| 46 | 3DShot2 | 18.38 | 15.29 | 21.94 | 31.11 | 26.07 | 18.12 | 29.17 | 23.10 | 30.44 | 0.49 | 0.38 | 0.57 | 0.36 | 18.38 | 15.29 | 21.94 | 31.11 | 26.07 | 18.12 | 29.17 | 23.10 | 30.44 | 0.01 | 0.02 | 3DShot2 | 46 | ||||||||||||
| 47 | Jones−UCL | 18.12 | 15.46 | 40.29 | 35.47 | 30.81 | 52.16 | 29.61 | 24.93 | 42.87 | 0.40 | 0.45 | 1.28 | 0.55 | 0.60 | 1.43 | 0.64 | 0.69 | 1.15 | 18.50 | 16.50 | 40.29 | 36.34 | 31.88 | 52.16 | 29.61 | 25.22 | 43.08 | 0.05 | 0.27 | 1.16 | 0.25 | 0.35 | 1.35 | 0.09 | 0.14 | 0.87 | Jones−UCL | 47 |
| 48 | TJ_Jiang | 18.00 | 13.58 | 41.04 | 33.43 | 29.17 | 51.35 | 30.70 | 22.95 | 43.60 | 0.36 | 1.35 | 0.20 | 0.28 | 1.38 | 0.79 | 0.33 | 1.23 | 21.25 | 16.58 | 41.20 | 40.70 | 30.62 | 51.35 | 30.70 | 24.64 | 44.21 | 0.96 | 0.30 | 1.26 | 1.05 | 0.08 | 1.29 | 0.24 | 0.05 | 1.00 | TJ_Jiang | 48 | |
| 49 | Zhang−Server | 17.88 | 13.79 | 38.22 | 37.50 | 31.11 | 48.79 | 28.95 | 21.64 | 41.15 | 0.32 | 1.09 | 0.90 | 0.66 | 1.21 | 0.54 | 0.09 | 0.96 | 20.88 | 18.92 | 38.22 | 40.99 | 34.98 | 52.27 | 35.09 | 31.43 | 41.15 | 0.84 | 1.20 | 0.95 | 1.10 | 1.02 | 1.36 | 0.86 | 1.20 | 0.64 | Zhang−Server | 49 | |
| 50 | keasar | 17.62 | 15.79 | 37.45 | 34.01 | 29.55 | 47.37 | 23.68 | 21.56 | 40.54 | 0.24 | 0.59 | 1.02 | 0.30 | 0.36 | 1.11 | 0.08 | 0.90 | 21.75 | 19.58 | 43.43 | 44.48 | 38.47 | 48.89 | 36.84 | 32.02 | 49.59 | 1.12 | 1.45 | 1.48 | 1.74 | 1.76 | 1.10 | 1.10 | 1.30 | 1.64 | keasar | 50 | |
| 51 | MUFOLD−MD | 17.62 | 15.29 | 38.39 | 40.12 | 34.69 | 44.54 | 21.71 | 20.47 | 43.63 | 0.24 | 0.38 | 1.11 | 1.35 | 1.37 | 0.93 | 1.23 | 20.12 | 19.75 | 41.03 | 45.35 | 44.38 | 49.17 | 30.26 | 22.81 | 45.54 | 0.59 | 1.51 | 1.24 | 1.90 | 3.03 | 1.12 | 0.18 | 1.16 | MUFOLD−MD | 51 | |||
| 52 | COMA | 17.62 | 14.38 | 7.87 | 40.99 | 32.46 | 18.60 | 12.50 | 9.43 | 7.93 | 0.24 | 0.02 | 1.50 | 0.93 | 17.62 | 14.38 | 17.03 | 40.99 | 32.46 | 24.68 | 19.08 | 16.01 | 22.29 | 1.10 | 0.48 | COMA | 52 | ||||||||||||
| 53 | 3DShotMQ | 17.62 | 14.29 | 24.08 | 31.98 | 22.87 | 23.77 | 28.29 | 22.30 | 31.26 | 0.24 | 0.45 | 0.21 | 17.62 | 14.29 | 24.08 | 31.98 | 22.87 | 23.77 | 28.29 | 22.30 | 31.26 | 3DShotMQ | 53 | |||||||||||||||
| 54 | COMA−M | 17.50 | 14.17 | 17.03 | 40.99 | 32.46 | 17.57 | 19.08 | 16.01 | 22.29 | 0.20 | 1.50 | 0.93 | 17.62 | 14.17 | 17.03 | 40.99 | 32.46 | 24.68 | 19.08 | 16.01 | 22.29 | 1.10 | 0.48 | COMA−M | 54 | |||||||||||||
| 55 | fais−server | 17.38 | 15.83 | 31.03 | 34.30 | 31.11 | 40.53 | 23.90 | 19.08 | 33.75 | 0.16 | 0.60 | 0.44 | 0.35 | 0.66 | 0.67 | 0.15 | 19.12 | 15.83 | 35.88 | 43.31 | 33.33 | 47.48 | 24.56 | 23.10 | 39.09 | 0.26 | 0.01 | 0.71 | 1.52 | 0.66 | 0.99 | 0.40 | fais−server | 55 | ||||
| 56 | A−TASSER | 17.38 | 15.83 | 33.61 | 37.50 | 33.43 | 43.49 | 26.32 | 23.83 | 36.35 | 0.16 | 0.60 | 0.68 | 0.90 | 1.12 | 0.86 | 0.17 | 0.49 | 0.44 | 22.50 | 19.38 | 36.66 | 50.00 | 35.76 | 43.94 | 37.94 | 33.55 | 42.55 | 1.37 | 1.37 | 0.79 | 2.75 | 1.18 | 0.72 | 1.26 | 1.56 | 0.80 | A−TASSER | 56 |
| 57 | HHpred2 | 17.38 | 15.25 | 19.05 | 38.95 | 33.91 | 20.04 | 20.39 | 16.74 | 23.35 | 0.16 | 0.37 | 1.15 | 1.21 | 17.38 | 15.25 | 19.05 | 38.95 | 33.91 | 20.04 | 20.39 | 16.74 | 23.35 | 0.73 | 0.79 | HHpred2 | 57 | ||||||||||||
| 58 | ZicoFullSTPFullData | 17.25 | 15.88 | 37.61 | 33.72 | 30.43 | 46.91 | 23.68 | 21.56 | 41.00 | 0.11 | 0.62 | 1.04 | 0.25 | 0.53 | 1.08 | 0.08 | 0.95 | 21.00 | 19.25 | 42.10 | 41.86 | 37.21 | 46.91 | 36.18 | 30.56 | 48.65 | 0.88 | 1.32 | 1.35 | 1.26 | 1.49 | 0.95 | 1.01 | 1.05 | 1.52 | ZicoFullSTPFullData | 58 | |
| 59 | POEM | 17.12 | 15.04 | 38.18 | 35.47 | 30.81 | 43.70 | 23.25 | 20.25 | 42.37 | 0.07 | 0.28 | 1.09 | 0.55 | 0.60 | 0.87 | 1.10 | 17.88 | 15.83 | 39.65 | 37.21 | 32.95 | 52.09 | 29.82 | 25.66 | 42.98 | 0.01 | 1.10 | 0.41 | 0.58 | 1.35 | 0.12 | 0.22 | 0.86 | POEM | 59 | |||
| 60 | GeneSilico | 17.00 | 15.42 | 34.63 | 31.11 | 25.10 | 44.03 | 29.82 | 27.05 | 37.83 | 0.03 | 0.44 | 0.77 | 0.90 | 0.67 | 1.07 | 0.60 | 19.25 | 16.25 | 38.29 | 34.01 | 29.36 | 44.03 | 33.77 | 28.07 | 44.66 | 0.30 | 0.17 | 0.96 | 0.73 | 0.67 | 0.63 | 1.05 | GeneSilico | 60 | ||||
| 61 | huber−torda−server | 16.88 | 14.46 | 14.70 | 35.47 | 28.49 | 31.27 | 18.42 | 15.50 | 11.48 | 0.05 | 0.55 | 0.15 | 0.06 | 16.88 | 14.46 | 14.70 | 35.47 | 28.49 | 31.27 | 22.59 | 17.76 | 15.61 | 0.09 | huber−torda−server | 61 | |||||||||||||
| 62 | Kolinski | 16.62 | 13.54 | 28.30 | 35.76 | 29.17 | 43.56 | 19.30 | 18.35 | 27.72 | 0.20 | 0.60 | 0.28 | 0.87 | 16.62 | 14.12 | 32.04 | 35.76 | 29.17 | 43.56 | 23.90 | 19.30 | 37.86 | 0.32 | 0.14 | 0.69 | 0.25 | Kolinski | 62 | ||||||||||
| 63 | Distill | 16.62 | 13.54 | 30.14 | 31.98 | 16.67 | 44.69 | 21.49 | 19.52 | 29.04 | 0.36 | 0.94 | 17.12 | 16.00 | 31.89 | 34.01 | 24.61 | 47.45 | 21.71 | 19.52 | 30.93 | 0.07 | 0.30 | 0.99 | Distill | 63 | |||||||||||||
| 64 | MidwayFolding | 16.25 | 14.04 | 30.68 | 33.72 | 28.49 | 43.89 | 18.42 | 15.06 | 30.07 | 0.41 | 0.25 | 0.15 | 0.89 | 16.38 | 14.04 | 32.85 | 33.72 | 28.49 | 43.89 | 24.34 | 20.69 | 34.94 | 0.40 | 0.72 | MidwayFolding | 64 | ||||||||||||
| 65 | RAPTOR | 16.25 | 13.58 | 38.34 | 38.08 | 31.49 | 48.37 | 23.90 | 21.71 | 41.56 | 1.10 | 1.00 | 0.74 | 1.18 | 0.11 | 1.01 | 18.38 | 13.58 | 39.57 | 38.08 | 31.49 | 48.37 | 26.97 | 23.90 | 44.70 | 0.01 | 1.09 | 0.57 | 0.27 | 1.06 | 1.06 | RAPTOR | 65 | ||||||
| 66 | fleil | 16.12 | 13.79 | 18.07 | 30.81 | 25.97 | 20.07 | 17.11 | 14.04 | 23.09 | 16.12 | 15.04 | 18.66 | 31.69 | 30.72 | 21.90 | 18.42 | 15.06 | 24.18 | 0.11 | fleil | 66 | |||||||||||||||||
| 67 | Poing | 16.00 | 13.25 | 23.49 | 27.91 | 26.55 | 29.86 | 28.07 | 23.25 | 27.02 | 0.42 | 0.38 | 21.00 | 17.42 | 27.16 | 29.36 | 26.65 | 33.38 | 31.36 | 26.39 | 30.89 | 0.88 | 0.62 | 0.33 | 0.34 | Poing | 67 | ||||||||||||
| 68 | FrankensteinLong | 15.88 | 13.67 | 29.10 | 30.23 | 27.33 | 21.89 | 26.54 | 22.66 | 40.05 | 0.27 | 0.20 | 0.28 | 0.84 | 15.88 | 14.04 | 29.10 | 33.72 | 30.52 | 29.41 | 26.54 | 22.66 | 40.05 | 0.02 | 0.06 | 0.51 | FrankensteinLong | 68 | |||||||||||
| 69 | mGenTHREADER | 15.88 | 12.62 | 23.95 | 30.52 | 26.45 | 20.35 | 28.07 | 23.39 | 32.47 | 0.42 | 0.41 | 0.01 | 15.88 | 12.62 | 23.95 | 30.52 | 26.45 | 20.35 | 28.07 | 23.39 | 32.47 | mGenTHREADER | 69 | |||||||||||||||
| 70 | SHORTLE | 15.75 | 14.25 | 32.15 | 32.56 | 29.07 | 38.66 | 22.59 | 21.13 | 34.60 | 0.54 | 0.05 | 0.26 | 0.55 | 0.00 | 0.25 | 15.75 | 14.25 | 32.15 | 35.47 | 33.14 | 43.42 | 22.59 | 21.13 | 34.60 | 0.33 | 0.09 | 0.62 | 0.68 | SHORTLE | 70 | ||||||||
| 71 | EB_AMU_Physics | 15.75 | 13.62 | 22.73 | 27.04 | 22.48 | 19.81 | 25.88 | 22.30 | 30.92 | 0.10 | 0.21 | 15.75 | 13.62 | 22.73 | 27.04 | 22.48 | 19.81 | 25.88 | 22.30 | 30.92 | EB_AMU_Physics | 71 | ||||||||||||||||
| 72 | Phragment | 15.62 | 12.67 | 23.37 | 25.87 | 22.77 | 17.58 | 26.32 | 21.34 | 31.49 | 0.17 | 0.04 | 15.75 | 13.42 | 25.47 | 27.91 | 22.77 | 29.47 | 26.75 | 22.66 | 31.49 | Phragment | 72 | ||||||||||||||||
| 73 | Sasaki−Cetin−Sasai | 15.62 | 11.62 | 39.24 | 34.30 | 27.52 | 39.13 | 21.49 | 21.34 | 47.27 | 1.18 | 0.35 | 0.58 | 0.04 | 1.63 | 16.25 | 15.54 | 41.55 | 34.88 | 33.14 | 45.30 | 25.22 | 23.61 | 48.05 | 1.29 | 0.62 | 0.83 | 1.45 | Sasaki−Cetin−Sasai | 73 | |||||||||
| 74 | FEIG | 15.50 | 12.83 | 18.27 | 28.78 | 26.07 | 22.58 | 16.67 | 13.74 | 21.68 | 15.50 | 13.46 | 25.44 | 29.65 | 26.36 | 35.24 | 22.15 | 17.47 | 26.99 | 0.05 | FEIG | 74 | |||||||||||||||||
| 75 | POEMQA | 15.38 | 13.46 | 39.28 | 30.52 | 27.81 | 48.13 | 25.44 | 21.93 | 42.98 | 1.19 | 0.01 | 1.16 | 0.04 | 0.15 | 1.16 | 23.12 | 20.54 | 39.65 | 36.34 | 29.84 | 52.09 | 39.91 | 35.38 | 42.98 | 1.58 | 1.82 | 1.10 | 0.25 | 1.35 | 1.53 | 1.87 | 0.86 | POEMQA | 75 | ||||
| 76 | Pcons_multi | 15.25 | 13.75 | 26.14 | 32.56 | 29.07 | 31.62 | 19.30 | 18.06 | 30.01 | 0.00 | 0.05 | 0.26 | 0.09 | 15.25 | 13.75 | 26.22 | 32.56 | 29.07 | 32.21 | 19.30 | 18.06 | 30.01 | Pcons_multi | 76 | ||||||||||||||
| 77 | PRI−Yang−KiharA | 15.25 | 13.67 | 26.55 | 32.56 | 28.88 | 33.33 | 19.30 | 18.35 | 29.84 | 0.04 | 0.05 | 0.22 | 0.20 | 15.25 | 13.67 | 26.55 | 32.56 | 28.88 | 33.33 | 19.30 | 18.35 | 29.84 | PRI−Yang−KiharA | 77 | ||||||||||||||
| 78 | MULTICOM | 15.25 | 13.62 | 38.97 | 30.23 | 27.52 | 48.15 | 25.66 | 22.15 | 42.67 | 1.16 | 1.17 | 0.07 | 0.19 | 1.13 | 20.88 | 19.04 | 42.18 | 41.86 | 36.82 | 48.15 | 36.18 | 30.26 | 48.74 | 0.84 | 1.24 | 1.36 | 1.26 | 1.41 | 1.04 | 1.01 | 1.00 | 1.54 | MULTICOM | 78 | ||||
| 79 | Sternberg | 15.25 | 12.50 | 13.32 | 33.72 | 28.39 | 36.11 | 26.10 | 21.86 | 7.82 | 0.25 | 0.13 | 0.38 | 0.13 | 0.13 | 22.88 | 21.46 | 39.57 | 35.76 | 28.39 | 52.11 | 39.69 | 37.21 | 41.68 | 1.50 | 2.17 | 1.09 | 0.14 | 1.35 | 1.50 | 2.18 | 0.70 | Sternberg | 79 | |||||
| 80 | AMU−Biology | 15.25 | 11.42 | 34.89 | 32.27 | 27.23 | 40.57 | 24.34 | 19.37 | 36.79 | 0.79 | 0.67 | 0.49 | 21.12 | 16.79 | 37.13 | 32.56 | 30.81 | 46.74 | 38.16 | 30.56 | 44.20 | 0.92 | 0.38 | 0.84 | 0.13 | 0.94 | 1.29 | 1.05 | 1.00 | AMU−Biology | 80 | |||||||
| 81 | Pcons_local | 14.88 | 14.12 | 27.36 | 31.39 | 29.65 | 34.55 | 19.52 | 17.18 | 30.76 | 0.11 | 0.38 | 0.28 | 15.50 | 14.12 | 30.62 | 31.39 | 29.65 | 34.55 | 27.41 | 22.30 | 40.09 | 0.17 | 0.00 | 0.51 | Pcons_local | 81 | ||||||||||||
| 82 | Pcons_dot_net | 14.88 | 13.29 | 26.22 | 31.69 | 27.62 | 32.21 | 19.08 | 17.69 | 29.83 | 0.01 | 0.12 | 15.75 | 14.12 | 27.36 | 34.30 | 29.65 | 34.55 | 26.32 | 23.54 | 30.76 | 0.00 | Pcons_dot_net | 82 | |||||||||||||||
| 83 | MUSTER | 14.62 | 13.46 | 28.28 | 29.94 | 25.97 | 34.14 | 23.25 | 18.27 | 32.01 | 0.20 | 0.25 | 18.88 | 15.00 | 28.98 | 36.34 | 31.01 | 38.34 | 23.25 | 20.32 | 32.01 | 0.18 | 0.00 | 0.25 | 0.17 | 0.29 | MUSTER | 83 | |||||||||||
| 84 | Phyre_de_novo | 14.38 | 12.88 | 19.64 | 22.96 | 18.80 | 11.62 | 25.22 | 22.59 | 30.08 | 0.01 | 0.27 | 16.12 | 13.12 | 28.40 | 26.16 | 24.23 | 23.38 | 28.29 | 23.03 | 37.93 | 0.26 | Phyre_de_novo | 84 | |||||||||||||||
| 85 | SMEG−CCP | 14.25 | 9.96 | 23.38 | 32.85 | 23.26 | 26.14 | 17.32 | 14.62 | 28.82 | 0.10 | 14.25 | 9.96 | 23.38 | 32.85 | 23.26 | 26.14 | 17.32 | 14.62 | 28.82 | SMEG−CCP | 85 | |||||||||||||||||
| 86 | BioSerf | 14.12 | 13.12 | 36.85 | 31.69 | 27.62 | 46.27 | 23.03 | 21.34 | 40.09 | 0.97 | 1.04 | 0.04 | 0.85 | 14.12 | 13.12 | 36.85 | 31.69 | 27.62 | 46.27 | 23.03 | 21.34 | 40.09 | 0.81 | 0.90 | 0.51 | BioSerf | 86 | |||||||||||
| 87 | Frankenstein | 14.00 | 10.21 | 19.91 | 30.81 | 22.09 | 27.50 | 18.64 | 15.72 | 21.69 | 15.38 | 13.92 | 29.39 | 33.14 | 30.33 | 34.70 | 26.10 | 21.71 | 40.63 | 0.05 | 0.02 | 0.01 | 0.58 | Frankenstein | 87 | ||||||||||||||
| 88 | FOLDpro | 13.88 | 12.29 | 6.53 | 30.52 | 27.52 | 3.94 | 14.69 | 12.43 | 12.63 | 13.88 | 12.29 | 13.49 | 30.52 | 27.52 | 14.96 | 17.11 | 15.94 | 18.22 | FOLDpro | 88 | ||||||||||||||||||
| 89 | Hao_Kihara | 13.88 | 11.62 | 22.59 | 31.98 | 27.62 | 12.40 | 22.59 | 17.25 | 34.51 | 0.24 | 16.88 | 14.33 | 22.59 | 33.14 | 27.62 | 19.21 | 22.59 | 17.25 | 34.51 | Hao_Kihara | 89 | |||||||||||||||||
| 90 | SAMUDRALA | 13.75 | 13.25 | 19.16 | 30.81 | 23.64 | 32.03 | 14.91 | 13.30 | 18.75 | 0.11 | 16.75 | 14.96 | 23.27 | 35.17 | 30.33 | 34.38 | 17.54 | 16.81 | 24.69 | 0.03 | 0.02 | SAMUDRALA | 90 | |||||||||||||||
| 91 | SAM−T06−server | 13.62 | 12.21 | 28.72 | 30.23 | 26.94 | 35.75 | 22.37 | 17.11 | 32.26 | 0.24 | 0.36 | 14.00 | 13.67 | 28.72 | 30.23 | 26.94 | 35.75 | 24.12 | 19.08 | 32.26 | 0.09 | SAM−T06−server | 91 | |||||||||||||||
| 92 | CpHModels | 13.62 | 12.12 | 31.69 | 28.20 | 27.15 | 0.00 | 0.00 | 0.09 | 13.62 | 12.12 | 31.69 | 28.20 | 27.15 | 0.00 | 0.00 | CpHModels | 92 | |||||||||||||||||||||
| 93 | Scheraga | 13.50 | 12.04 | 22.64 | 27.91 | 21.32 | 30.30 | 18.20 | 14.40 | 25.41 | 15.12 | 13.96 | 25.77 | 32.56 | 29.75 | 33.91 | 23.46 | 19.96 | 31.50 | Scheraga | 93 | ||||||||||||||||||
| 94 | GS−KudlatyPred | 13.50 | 11.12 | 17.75 | 30.52 | 24.81 | 28.59 | 20.61 | 19.01 | 18.39 | 14.88 | 13.29 | 17.75 | 32.27 | 28.00 | 28.59 | 24.78 | 22.00 | 18.88 | GS−KudlatyPred | 94 | ||||||||||||||||||
| 95 | 3Dpro | 13.25 | 11.92 | 19.64 | 26.45 | 20.83 | 15.93 | 23.68 | 19.74 | 27.89 | 15.38 | 12.96 | 19.64 | 31.39 | 28.68 | 19.91 | 23.68 | 19.74 | 27.89 | 3Dpro | 95 | ||||||||||||||||||
| 96 | pipe_int | 13.12 | 11.38 | 23.93 | 27.91 | 26.26 | 19.07 | 19.08 | 16.52 | 32.33 | 13.12 | 11.38 | 23.93 | 27.91 | 26.26 | 19.07 | 19.08 | 16.52 | 32.33 | pipe_int | 96 | ||||||||||||||||||
| 97 | RANDOM | 12.96 | 10.74 | 12.96 | 28.13 | 24.39 | 28.13 | 16.42 | 14.10 | 16.42 | 12.96 | 10.74 | 12.96 | 28.13 | 24.39 | 28.13 | 16.42 | 14.10 | 16.42 | RANDOM | 97 | ||||||||||||||||||
| 98 | POISE | 12.88 | 12.46 | 31.90 | 26.74 | 25.77 | 36.38 | 20.83 | 19.08 | 37.03 | 0.52 | 0.40 | 0.51 | 16.12 | 13.71 | 31.90 | 31.39 | 28.68 | 36.38 | 25.44 | 20.76 | 37.03 | 0.30 | 0.14 | 0.15 | POISE | 98 | ||||||||||||
| 99 | LEE | 12.88 | 10.71 | 23.93 | 29.65 | 24.42 | 28.18 | 20.39 | 17.76 | 28.74 | 14.25 | 12.25 | 24.74 | 29.65 | 25.97 | 30.69 | 24.78 | 21.27 | 28.81 | LEE | 99 | ||||||||||||||||||
| 100 | CBSU | 12.88 | 10.67 | 20.78 | 29.07 | 23.93 | 19.80 | 18.42 | 17.84 | 27.73 | 12.88 | 10.67 | 20.78 | 29.07 | 23.93 | 19.80 | 18.42 | 17.84 | 27.73 | CBSU | 100 | ||||||||||||||||||
| 101 | tripos_08 | 12.75 | 10.33 | 10.21 | 26.45 | 22.19 | 7.94 | 15.13 | 13.09 | 16.60 | 12.75 | 10.33 | 10.87 | 26.45 | 22.19 | 7.94 | 16.67 | 15.57 | 17.67 | tripos_08 | 101 | ||||||||||||||||||
| 102 | Softberry | 12.50 | 11.12 | 28.18 | 24.71 | 23.45 | 31.69 | 19.52 | 16.45 | 33.63 | 0.19 | 0.09 | 0.14 | 12.50 | 11.12 | 28.18 | 24.71 | 23.45 | 31.69 | 19.52 | 16.45 | 33.63 | Softberry | 102 | |||||||||||||||
| 103 | forecast | 12.50 | 10.21 | 19.04 | 28.20 | 23.45 | 25.97 | 20.18 | 16.23 | 21.14 | 13.50 | 11.21 | 19.65 | 28.20 | 23.84 | 26.49 | 20.18 | 17.40 | 22.83 | forecast | 103 | ||||||||||||||||||
| 104 | DistillSN | 12.38 | 11.75 | 17.16 | 26.74 | 25.19 | 14.98 | 19.74 | 15.64 | 24.22 | 12.88 | 11.75 | 17.16 | 26.74 | 25.19 | 14.98 | 20.61 | 18.06 | 25.81 | DistillSN | 104 | ||||||||||||||||||
| 105 | StruPPi | 11.88 | 8.58 | 6.04 | 16.28 | 13.57 | 20.18 | 13.30 | 24.74 | 11.88 | 8.58 | 6.04 | 16.28 | 13.57 | 20.18 | 13.30 | 24.74 | StruPPi | 105 | ||||||||||||||||||||
| 106 | 3D−JIGSAW_AEP | 11.75 | 10.71 | 27.33 | 24.90 | 18.74 | 0.00 | 0.00 | 12.12 | 11.42 | 28.20 | 26.55 | 20.12 | 0.00 | 0.00 | 3D−JIGSAW_AEP | 106 | ||||||||||||||||||||||
| 107 | ProtAnG | 11.62 | 9.92 | 14.19 | 22.96 | 19.09 | 19.30 | 17.98 | 28.80 | 11.62 | 9.92 | 14.19 | 22.96 | 19.09 | 19.30 | 17.98 | 28.80 | ProtAnG | 107 | ||||||||||||||||||||
| 108 | psiphifoldings | 11.50 | 11.21 | 21.27 | 25.29 | 24.61 | 22.86 | 14.69 | 12.21 | 26.90 | 11.50 | 11.21 | 21.27 | 25.29 | 24.61 | 22.86 | 14.69 | 12.21 | 26.90 | psiphifoldings | 108 | ||||||||||||||||||
| 109 | ACOMPMOD | 11.50 | 11.00 | 16.08 | 25.00 | 24.03 | 7.92 | 19.52 | 18.64 | 26.14 | 14.25 | 12.83 | 16.50 | 29.65 | 25.39 | 22.68 | 21.71 | 18.64 | 26.14 | ACOMPMOD | 109 | ||||||||||||||||||
| 110 | FUGUE_KM | 11.12 | 9.67 | 9.25 | 24.13 | 23.35 | 6.91 | 16.67 | 15.06 | 15.40 | 13.62 | 13.04 | 9.69 | 27.62 | 24.71 | 10.69 | 20.61 | 16.08 | 16.63 | FUGUE_KM | 110 | ||||||||||||||||||
| 111 | SAM−T02−server | 11.12 | 9.54 | 25.87 | 22.38 | 0.00 | 0.00 | 11.50 | 10.17 | 26.74 | 23.64 | 0.00 | 0.00 | SAM−T02−server | 111 | ||||||||||||||||||||||||
| 112 | PS2−server | 11.00 | 9.33 | 2.31 | 25.58 | 21.61 | 24.61 | 12.28 | 10.53 | 15.38 | 13.00 | 27.03 | 32.56 | 27.23 | 43.41 | 26.54 | 20.83 | 30.64 | 0.68 | PS2−server | 112 | ||||||||||||||||||
| 113 | PS2−manual | 11.00 | 9.33 | 2.31 | 25.58 | 21.61 | 24.61 | 12.28 | 10.53 | 15.38 | 13.00 | 27.03 | 32.56 | 27.23 | 43.41 | 26.54 | 20.83 | 35.26 | 0.68 | PS2−manual | 113 | ||||||||||||||||||
| 114 | nFOLD3 | 10.88 | 9.54 | 26.87 | 24.42 | 20.54 | 34.41 | 16.23 | 14.91 | 30.15 | 0.07 | 0.27 | 18.25 | 14.29 | 27.84 | 36.34 | 26.55 | 34.41 | 24.78 | 20.39 | 33.37 | 0.25 | nFOLD3 | 114 | |||||||||||||||
| 115 | TWPPLAB | 10.75 | 10.04 | 9.77 | 23.84 | 21.90 | 16.23 | 13.45 | 20.40 | 10.75 | 10.04 | 9.77 | 23.84 | 21.90 | 16.23 | 13.45 | 20.40 | TWPPLAB | 115 | ||||||||||||||||||||
| 116 | panther_server | 10.75 | 8.79 | 17.73 | 13.47 | 15.79 | 12.13 | 3.43 | 10.75 | 8.79 | 17.73 | 13.47 | 15.79 | 12.13 | 3.43 | panther_server | 116 | ||||||||||||||||||||||
| 117 | DelCLab | 10.38 | 8.71 | 12.38 | 22.09 | 20.06 | 26.64 | 14.47 | 11.40 | 10.65 | 11.88 | 10.25 | 12.38 | 26.45 | 23.06 | 26.64 | 14.47 | 12.28 | 15.28 | DelCLab | 117 | ||||||||||||||||||
| 118 | 3D−JIGSAW_V3 | 10.00 | 9.12 | 23.26 | 21.22 | 17.67 | 0.00 | 0.00 | 12.25 | 9.46 | 28.49 | 22.00 | 24.02 | 0.00 | 0.00 | 3D−JIGSAW_V3 | 118 | ||||||||||||||||||||||
| 119 | OLGAFS | 9.88 | 8.58 | 0.00 | 0.00 | 17.32 | 15.06 | 0.48 | 11.62 | 9.88 | 24.71 | 20.83 | 17.32 | 15.06 | 0.48 | OLGAFS | 119 | ||||||||||||||||||||||
| 120 | mariner1 | 9.88 | 7.96 | 8.77 | 20.93 | 16.38 | 16.45 | 13.09 | 18.05 | 19.88 | 17.29 | 31.69 | 28.20 | 24.23 | 26.55 | 35.09 | 30.56 | 42.69 | 0.51 | 0.57 | 0.28 | 0.86 | 1.05 | 0.82 | mariner1 | 120 | |||||||||||||
| 121 | keasar−server | 9.75 | 8.17 | 17.15 | 15.70 | 13.16 | 11.84 | 12.48 | 18.12 | 14.50 | 15.54 | 28.78 | 26.16 | 19.16 | 32.02 | 25.66 | 35.96 | 0.42 | 0.22 | 0.03 | keasar−server | 121 | |||||||||||||||||
| 122 | schenk−torda−server | 9.62 | 8.29 | 8.41 | 20.64 | 17.54 | 13.82 | 12.35 | 21.31 | 9.88 | 8.62 | 8.41 | 20.64 | 17.54 | 17.54 | 15.21 | 21.33 | schenk−torda−server | 122 | ||||||||||||||||||||
| 123 | rehtnap | 8.00 | 7.33 | 0.00 | 0.00 | 14.04 | 12.87 | 8.00 | 7.67 | 0.00 | 0.00 | 14.04 | 13.45 | rehtnap | 123 | ||||||||||||||||||||||||
| 124 | Pushchino | 6.12 | 5.62 | 0.00 | 0.00 | 10.75 | 9.87 | 6.12 | 5.62 | 0.00 | 0.00 | 10.75 | 9.87 | Pushchino | 124 | ||||||||||||||||||||||||
| 125 | Abagyan | Abagyan | 125 | ||||||||||||||||||||||||||||||||||||
| 126 | BHAGEERATH | BHAGEERATH | 126 | ||||||||||||||||||||||||||||||||||||
| 127 | FEIG_REFINE | FEIG_REFINE | 127 | ||||||||||||||||||||||||||||||||||||
| 128 | FLOUDAS | FLOUDAS | 128 | ||||||||||||||||||||||||||||||||||||
| 129 | Fiser−M4T | Fiser−M4T | 129 | ||||||||||||||||||||||||||||||||||||
| 130 | HCA | HCA | 130 | ||||||||||||||||||||||||||||||||||||
| 131 | Handl−Lovell | Handl−Lovell | 131 | ||||||||||||||||||||||||||||||||||||
| 132 | JIVE08 | JIVE08 | 132 | ||||||||||||||||||||||||||||||||||||
| 133 | KudlatyPredHuman | KudlatyPredHuman | 133 | ||||||||||||||||||||||||||||||||||||
| 134 | LEE−SERVER | LEE−SERVER | 134 | ||||||||||||||||||||||||||||||||||||
| 135 | LOOPP_Server | LOOPP_Server | 135 | ||||||||||||||||||||||||||||||||||||
| 136 | Linnolt−UH−CMB | Linnolt−UH−CMB | 136 | ||||||||||||||||||||||||||||||||||||
| 137 | MeilerLabRene | MeilerLabRene | 137 | ||||||||||||||||||||||||||||||||||||
| 138 | NIM2 | NIM2 | 138 | ||||||||||||||||||||||||||||||||||||
| 139 | Nano_team | Nano_team | 139 | ||||||||||||||||||||||||||||||||||||
| 140 | NirBenTal | NirBenTal | 140 | ||||||||||||||||||||||||||||||||||||
| 141 | Ozkan−Shell | Ozkan−Shell | 141 | ||||||||||||||||||||||||||||||||||||
| 142 | PHAISTOS | PHAISTOS | 142 | ||||||||||||||||||||||||||||||||||||
| 143 | PZ−UAM | PZ−UAM | 143 | ||||||||||||||||||||||||||||||||||||
| 144 | ProteinShop | ProteinShop | 144 | ||||||||||||||||||||||||||||||||||||
| 145 | RPFM | RPFM | 145 | ||||||||||||||||||||||||||||||||||||
| 146 | ShakAbInitio | ShakAbInitio | 146 | ||||||||||||||||||||||||||||||||||||
| 147 | TsaiLab | TsaiLab | 147 | ||||||||||||||||||||||||||||||||||||
| 148 | UCDavisGenome | UCDavisGenome | 148 | ||||||||||||||||||||||||||||||||||||
| 149 | Wolfson−FOBIA | Wolfson−FOBIA | 149 | ||||||||||||||||||||||||||||||||||||
| 150 | Wolynes | Wolynes | 150 | ||||||||||||||||||||||||||||||||||||
| 151 | YASARA | YASARA | 151 | ||||||||||||||||||||||||||||||||||||
| 152 | YASARARefine | YASARARefine | 152 | ||||||||||||||||||||||||||||||||||||
| 153 | Zhou−SPARKS | Zhou−SPARKS | 153 | ||||||||||||||||||||||||||||||||||||
| 154 | dill_ucsf | dill_ucsf | 154 | ||||||||||||||||||||||||||||||||||||
| 155 | dill_ucsf_extended | dill_ucsf_extended | 155 | ||||||||||||||||||||||||||||||||||||
| 156 | igor | igor | 156 | ||||||||||||||||||||||||||||||||||||
| 157 | jacobson | jacobson | 157 | ||||||||||||||||||||||||||||||||||||
| 158 | mahmood−torda−server | mahmood−torda−server | 158 | ||||||||||||||||||||||||||||||||||||
| 159 | mti | mti | 159 | ||||||||||||||||||||||||||||||||||||
| 160 | mumssp | mumssp | 160 | ||||||||||||||||||||||||||||||||||||
| 161 | ricardo | ricardo | 161 | ||||||||||||||||||||||||||||||||||||
| 162 | rivilo | rivilo | 162 | ||||||||||||||||||||||||||||||||||||
| 163 | sessions | sessions | 163 | ||||||||||||||||||||||||||||||||||||
| 164 | taylor | taylor | 164 | ||||||||||||||||||||||||||||||||||||
| 165 | test_http_server_01 | test_http_server_01 | 165 | ||||||||||||||||||||||||||||||||||||
| 166 | xianmingpan | xianmingpan | 166 | ||||||||||||||||||||||||||||||||||||