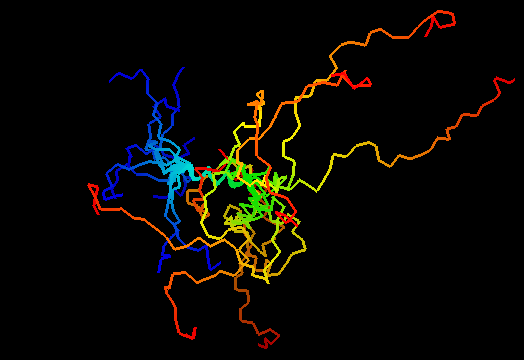
T0484
RhR2 Rhodobacter sphaeroides
Target sequence:
>T0484 RhR2, Rhodobacter sphaeroides, 62 residues
MRDMTEETRKDLPPEALRALAEAEERRRRAKALDLPKEIGGRNGPEPVRFGDWEKKGIAIDF
Structure:
Method: NMR
Determined by:
NESG
PDB ID: 2k5k
Domains: PyMOL of domains
Just a part of an α-helix is ordered in NMR models. This should be a two-domain protein, possibly forming dimers, but NMR models were too poor to reveal the structure of a very sequence-conserved short C-terminal domain. This target should probably be removed and predictions not evaluated.
Structure classification:
Unknown, as the C-terminal domain was unstructured.
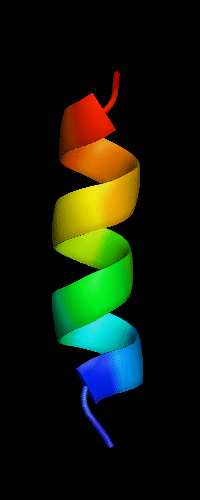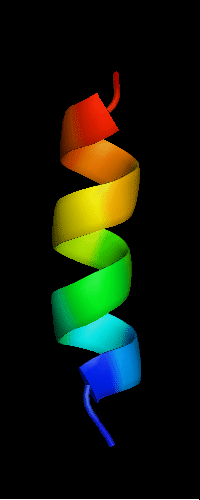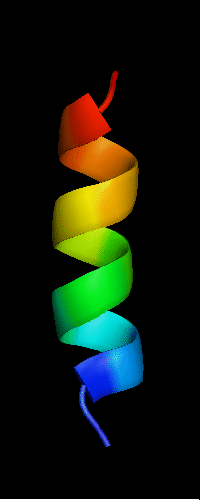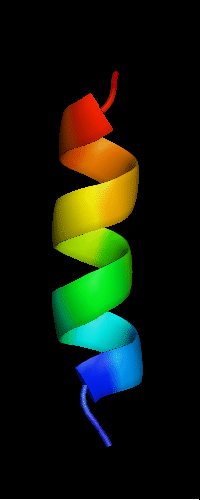
Cartoon diagram of 484:
2k5k chain 1 residues 13-26
CASP category:
Comparative modeling:easy. This is a joke, as many servers predicted the sinlge α-helix with 100% accuracy. This target should probably be removed and predictions not evaluated.
Closest templates:
Any α-helix will do for the target structure. No templates are available for the C-terminal domain, which appears disordered in the NMR models (it is strange!), and the C-terminal domain should be modeled de-novo:
Conservation: 6 59 6 7 88 8 88 T0484_RhR2 ALDL-PKEIGGR-NGPEPVRFG---DWEKKGIAIDF----- 62
gi_46203266 KIAE-RRETLGR-GGLEPVRYE---DWEVKGLASDF----- 79
gi_188579789 RIAE-RPESLGR-GGLEPVRYE---DWEVKGLASDF----- 75gi_156452939 KIAE-RPETLGR-GGLEPVRYE---DWEVKGLASDF----- 75gi_163701318 EIRR-AREIDGR-GGLEPVRYD---DWEVKGLATDF----- 68gi_94498067 PDPI-DNPTPHD-EAMSPVRYG---DWERKGIAIDF----- 57
gi_83950713 EAPL-PKELGGR-KGPEPVRYG---DWEKKGIAVDF----- 64gi_110678029 DMNM-PPELGGR-EGPEPVRYG---DWERKGIAVDF----- 64gi_71275730 QRSN-LEEVGGR-GGLDPVRYG---DWEKDGRCIDF----- 72
gi_114706036 PDSL-PSEVDGRKSGVEPVRYG---DWEKDGIVSDF----- 80
gi_146337495 AKPM-PKELQGP-KGPEPTRYG---DWENKGIISDF----- 63
gi_92115830 ASAG-PKEIQGP-KGPEPTRFG---DWERNGIASDF----- 72gi_27375689 AKAT-PKELQGP-KGPEPTRYG---DWERKGIASDF----- 69gi_115466524 VNKD-TGEIGGP-RGPEPTRYG---DWERGGRCSDF----- 122
gi_118483577 VNKE-TGEIGGP-KGPEPTRFG---DWERNGRCSDF----- 118
gi_116254450 PLEL-PPEIGGR-GGAEPARFG---DYEINGRAIDF----- 74
gi_190893966 PLEL-APEIGGR-GGAEPARFG---DYEINGRAIDF----- 74gi_163757450 KIST-EKEIGGR-GGADPARFG---DWEIDGRAIDF----- 79gi_188576490 KKPL-PKEIGGR-DGPEPTRYG---DWEKNGRCIDF----- 88
gi_21231577 KKPL-PKEIGGR-DGPEPTRYG---DWEKNGRCIDF----- 88gi_190615226 VNPH-TGEVGGP-AGPEPTRYG---DWERKGRVTDF----- 127
gi_190660756 TNPY-TGEIGGQ-AGPEPTRYG---DWERKGRVTDF----- 93gi_25151100 VNKS-TGEVGGP-AGPEPTRYG---DWERKGRVSDF----- 89
gi_56118809 INPV-TKEKGGP-RGPEPTRYG---DWERKGRCIDF----- 118
gi_118088802 INPA-TKERGGP-KGPEPTRFG---DWERKGRCIDF----- 92gi_162149216 KPAE-PHEYGGP-KEQRPTRYG---DWTVKGRCIDF----- 52
gi_159487193 PNPD-TGELYGP-RGREPTRYG---DWENKGKCIDF----- 151
gi_114797835 AAALPKDEFGGP-KSIEPTRYG---DWERKGIAYDF----- 69
gi_157828085 NLPK-EKEIGGV-KGLEPTRYG---DWQHKGKVTDF----- 94
gi_165932785 NLPK-EKEIGGV-KGLEPTRYG---DWQHKGKVTDF----- 64gi_157964234 NLPK-EKEIGGV-KGLEPTRYG---DWQYKGKVTDF----- 106gi_67459504 KLPK-EKEIGGV-KGLEPTRYG---DWQHKGKVTDF----- 78gi_159185403 DLQL-PPETGGR-GGAEPVRFG---DYEIKGRAIDF----- 70
gi_163869267 TKEQ-PLENGGR-GGKDPSRYG---DWEIKGRAIDF----- 71
gi_49474773 NKDK-PLENGGR-GGKDPSRYG---DWEIKGRAIDF----- 86gi_121602399 HEKK-PLESGGR-GGKDPARYG---DWEIKGRAIDF----- 68gi_116788769 VNSE-TGERGGP-RGPEPTRYG---DWEKGGRCSDF----- 129
gi_85375087 PQPI-DAEEKDP-DGLSPTRYG---DWVKDGVAIDF----- 55
gi_85710226 PKAV-DPMKDEP-REISPTRYG---DWEKNGIAWDF----- 66
gi_71656832 VDET-TGTPIGS-TQLSPTRYG---DWESNGRCHDF----- 107
gi_157864462 VDEE-TGRVIGS-TQPDPTRYG---DWEVNGRCYDF----- 185
gi_58267704 VNPK-TGEQGGP--KNDPFVAG-DADWQFGGRVTCVGMMMD 125
gi_149247609 VNPK-TGERGGP--KQDPLRNG--EEWTYNGRTIDF----- 177
gi_50405785 VNPV-TGERGGP--KQDPLKHG--DEWSFNGRVTDF----- 135
gi_190344418 VNPK-TGERGGP--KQDPLRHG--TDWSFNGRVTDF----- 125gi_115384010 KNPK-TGEIGGP--KNEPLRWGSEGDWSYGGRVTDF----- 139
gi_169774775 RNPK-TGEVGGP--KNEPLRWGAEGDWSYGGRVTDF----- 150gi_121715840 KNPK-TGEIGGP--KNEPLRWGAGGDWSYGGRVTDF----- 151gi_119190321 VNPK-TGEVGGP--KNEPLRWGSGGDWSYGGRVTDF----- 158gi_189198608 VNPK-TGEVGGP--KNEPLRWGGGGDWSYNGRVTDF----- 166gi_171677245 VNPK-TGEVGGP--KNEPLRWGDKGDWSYNGRVTDF----- 153
gi_46137775 VNPK-TGEVGGP--KNDPLRWGGESDWSYNGRATDF----- 137gi_85110151 KNPK-TGEIGGP--KNEPLRWGAGGDWSYNGRVTDF----- 169gi_50310269 KNPK-TGEIGGP--KQDPLRHG---DYSFNGRVTDF----- 131
gi_50543020 RNPK-TGEIGGP--KQDPLKHG---DYSFNGRCTDF----- 124gi_84999584 HPQI-GKEYGSVYTKHEPTMFG---DWSHMGRVTDF----- 185
gi_156086102 LKNG-AKEFGNVYKMHEPTMFG---DWSHMGRVSDF----- 131
gi_124805863 INNI-SEECGYKYDKHEPTMFG---DWSHNCRVTDF----- 140
gi_83286403 EKIM-SEEYGYKFDKHEPTMFG---DWSHNCRVTDF----- 137
Consensus_aa: .....s.E.st.....-PhR@G...DWp.pG.hhDF.....Consensus_ss: ee
Target sequence - PDB file inconsistencies:
None. PDB sequence matches target sequence exactly.
T0484 2k5k.pdb T0484.pdb PyMOL PyMOL of domains
T0484 1 MRDMTEETRKDLPPEALRALAEAEERRRRAKALDLPKEIGGRNGPEPVRFGDWEKKGIAIDF 62 ************||||||||||||||************************************ 2k5kA 1 MRDMTEETRKDLPPEALRALAEAEERRRRAKALDLPKEIGGRNGPEPVRFGDWEKKGIAIDF 62
Residue change log: remove 1-MRDMTEETRKDL-12, 27-RRRAKALDLPKEIGGRNGPEPVRFGDWEKKGIAIDF-62 by 3.5Å SD cutoff;
Single domain protein: target 13-26 ; pdb 13-26
Sequence classification:
Protein of unknown function (DUF1674) in Pfam. N-terminal sequence domain is a coiled-coil, C-terminal domain is very short, but very conserved, we term it WF-cage domain.
Comments:
Very insteresting protein. Can be a prototype for a large domain family, so its structure should be properly determined.
Server predictions:
T0484:pdb 13-26:seq 13-26:CM_easy; alignment
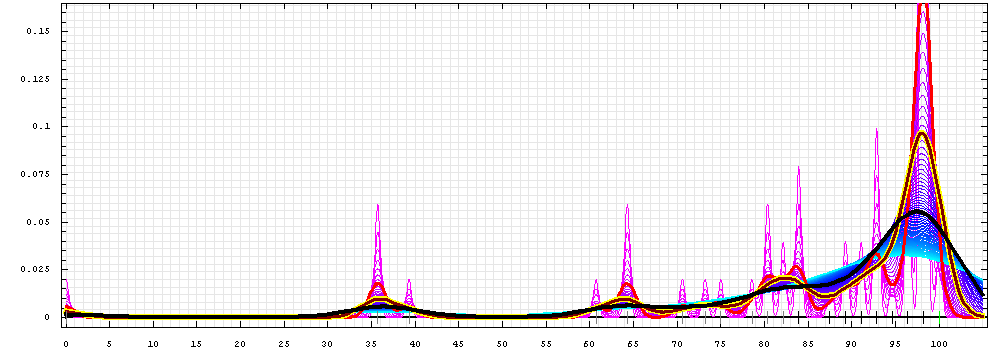
First models for T0484:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0484 | T0484 | T0484 | T0484 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | mufold | 100.00 | 100.00 | 96.68 | 0.90 | 0.81 | 0.86 | 100.00 | 100.00 | 96.70 | 0.83 | 0.76 | 0.69 | mufold | 1 |
| 2 | Elofsson | 100.00 | 100.00 | 94.31 | 0.90 | 0.81 | 0.51 | 100.00 | 100.00 | 95.53 | 0.83 | 0.76 | 0.46 | Elofsson | 2 |
| 3 | SAM−T06−server | 100.00 | 100.00 | 97.17 | 0.90 | 0.81 | 0.93 | 100.00 | 100.00 | 97.17 | 0.83 | 0.76 | 0.78 | SAM−T06−server | 3 |
| 4 | MUSTER | 100.00 | 100.00 | 96.57 | 0.90 | 0.81 | 0.84 | 100.00 | 100.00 | 96.57 | 0.83 | 0.76 | 0.66 | MUSTER | 4 |
| 5 | fams−ace2 | 98.21 | 98.21 | 93.94 | 0.63 | 0.61 | 0.46 | 98.21 | 98.21 | 95.34 | 0.46 | 0.46 | 0.42 | fams−ace2 | 5 |
| 6 | keasar | 98.21 | 98.21 | 95.36 | 0.63 | 0.61 | 0.66 | 98.21 | 98.21 | 96.31 | 0.46 | 0.46 | 0.61 | keasar | 6 |
| 7 | MULTICOM−RANK | 98.21 | 98.21 | 94.89 | 0.63 | 0.61 | 0.60 | 98.21 | 98.21 | 95.53 | 0.46 | 0.46 | 0.46 | MULTICOM−RANK | 7 |
| 8 | BAKER−ROBETTA | 98.21 | 98.21 | 95.71 | 0.63 | 0.61 | 0.72 | 98.21 | 98.21 | 95.71 | 0.46 | 0.46 | 0.50 | BAKER−ROBETTA | 8 |
| 9 | TASSER | 98.21 | 98.21 | 95.54 | 0.63 | 0.61 | 0.69 | 98.21 | 98.21 | 95.54 | 0.46 | 0.46 | 0.46 | TASSER | 9 |
| 10 | SAM−T08−human | 98.21 | 98.21 | 94.16 | 0.63 | 0.61 | 0.49 | 98.21 | 98.21 | 95.97 | 0.46 | 0.46 | 0.55 | SAM−T08−human | 10 |
| 11 | LevittGroup | 98.21 | 98.21 | 95.00 | 0.63 | 0.61 | 0.61 | 98.21 | 98.21 | 96.30 | 0.46 | 0.46 | 0.61 | LevittGroup | 11 |
| 12 | DBAKER | 98.21 | 98.21 | 96.08 | 0.63 | 0.61 | 0.77 | 98.21 | 98.21 | 96.34 | 0.46 | 0.46 | 0.62 | DBAKER | 12 |
| 13 | POEMQA | 98.21 | 98.21 | 96.22 | 0.63 | 0.61 | 0.79 | 98.21 | 98.21 | 96.22 | 0.46 | 0.46 | 0.60 | POEMQA | 13 |
| 14 | Zico | 98.21 | 98.21 | 95.47 | 0.63 | 0.61 | 0.68 | 98.21 | 98.21 | 96.22 | 0.46 | 0.46 | 0.60 | Zico | 14 |
| 15 | GS−MetaServer2 | 98.21 | 98.21 | 94.49 | 0.63 | 0.61 | 0.54 | 98.21 | 98.21 | 94.79 | 0.46 | 0.46 | 0.32 | GS−MetaServer2 | 15 |
| 16 | GeneSilicoMetaServer | 98.21 | 98.21 | 94.49 | 0.63 | 0.61 | 0.54 | 98.21 | 98.21 | 94.71 | 0.46 | 0.46 | 0.30 | GeneSilicoMetaServer | 16 |
| 17 | FLOUDAS | 98.21 | 98.21 | 95.98 | 0.63 | 0.61 | 0.76 | 98.21 | 98.21 | 95.98 | 0.46 | 0.46 | 0.55 | FLOUDAS | 17 |
| 18 | fleil | 98.21 | 98.21 | 96.21 | 0.63 | 0.61 | 0.79 | 98.21 | 98.21 | 96.21 | 0.46 | 0.46 | 0.59 | fleil | 18 |
| 19 | Sasaki−Cetin−Sasai | 98.21 | 98.21 | 92.10 | 0.63 | 0.61 | 0.18 | 98.21 | 98.21 | 92.38 | 0.46 | 0.46 | Sasaki−Cetin−Sasai | 19 | |
| 20 | GS−KudlatyPred | 98.21 | 98.21 | 94.30 | 0.63 | 0.61 | 0.51 | 98.21 | 98.21 | 95.48 | 0.46 | 0.46 | 0.45 | GS−KudlatyPred | 20 |
| 21 | keasar−server | 98.21 | 98.21 | 95.79 | 0.63 | 0.61 | 0.73 | 98.21 | 98.21 | 95.80 | 0.46 | 0.46 | 0.51 | keasar−server | 21 |
| 22 | Ozkan−Shell | 98.21 | 98.21 | 94.64 | 0.63 | 0.61 | 0.56 | 98.21 | 98.21 | 94.64 | 0.46 | 0.46 | 0.29 | Ozkan−Shell | 22 |
| 23 | METATASSER | 98.21 | 98.21 | 93.72 | 0.63 | 0.61 | 0.42 | 98.21 | 98.21 | 95.47 | 0.46 | 0.46 | 0.45 | METATASSER | 23 |
| 24 | SAINT1 | 98.21 | 98.21 | 94.48 | 0.63 | 0.61 | 0.53 | 98.21 | 98.21 | 94.48 | 0.46 | 0.46 | 0.25 | SAINT1 | 24 |
| 25 | SAM−T08−server | 98.21 | 98.21 | 95.81 | 0.63 | 0.61 | 0.73 | 98.21 | 98.21 | 95.81 | 0.46 | 0.46 | 0.52 | SAM−T08−server | 25 |
| 26 | LEE | 98.21 | 98.21 | 92.81 | 0.63 | 0.61 | 0.29 | 98.21 | 98.21 | 96.24 | 0.46 | 0.46 | 0.60 | LEE | 26 |
| 27 | Jones−UCL | 98.21 | 98.21 | 95.38 | 0.63 | 0.61 | 0.67 | 98.21 | 98.21 | 95.38 | 0.46 | 0.46 | 0.43 | Jones−UCL | 27 |
| 28 | MUFOLD−Server | 98.21 | 98.21 | 95.06 | 0.63 | 0.61 | 0.62 | 98.21 | 98.21 | 95.49 | 0.46 | 0.46 | 0.45 | MUFOLD−Server | 28 |
| 29 | Distill | 98.21 | 98.21 | 91.69 | 0.63 | 0.61 | 0.12 | 100.00 | 100.00 | 95.75 | 0.83 | 0.76 | 0.50 | Distill | 29 |
| 30 | OLGAFS | 98.21 | 98.21 | 94.79 | 0.63 | 0.61 | 0.58 | 98.21 | 98.21 | 95.93 | 0.46 | 0.46 | 0.54 | OLGAFS | 30 |
| 31 | FAMS−multi | 98.21 | 98.21 | 92.69 | 0.63 | 0.61 | 0.27 | 98.21 | 98.21 | 93.55 | 0.46 | 0.46 | 0.07 | FAMS−multi | 31 |
| 32 | AMU−Biology | 98.21 | 98.21 | 95.52 | 0.63 | 0.61 | 0.69 | 98.21 | 98.21 | 95.52 | 0.46 | 0.46 | 0.46 | AMU−Biology | 32 |
| 33 | Zhang | 98.21 | 98.21 | 95.84 | 0.63 | 0.61 | 0.74 | 98.21 | 98.21 | 95.87 | 0.46 | 0.46 | 0.53 | Zhang | 33 |
| 34 | xianmingpan | 98.21 | 98.21 | 92.99 | 0.63 | 0.61 | 0.31 | 98.21 | 98.21 | 92.99 | 0.46 | 0.46 | xianmingpan | 34 | |
| 35 | SHORTLE | 98.21 | 98.21 | 95.35 | 0.63 | 0.61 | 0.66 | 98.21 | 98.21 | 95.84 | 0.46 | 0.46 | 0.52 | SHORTLE | 35 |
| 36 | McGuffin | 98.21 | 98.21 | 95.48 | 0.63 | 0.61 | 0.68 | 98.21 | 98.21 | 95.55 | 0.46 | 0.46 | 0.46 | McGuffin | 36 |
| 37 | Phyre2 | 98.21 | 98.21 | 95.97 | 0.63 | 0.61 | 0.75 | 98.21 | 98.21 | 95.97 | 0.46 | 0.46 | 0.55 | Phyre2 | 37 |
| 38 | RBO−Proteus | 98.21 | 98.21 | 95.63 | 0.63 | 0.61 | 0.70 | 98.21 | 98.21 | 96.34 | 0.46 | 0.46 | 0.62 | RBO−Proteus | 38 |
| 39 | FALCON | 98.21 | 98.21 | 95.48 | 0.63 | 0.61 | 0.68 | 98.21 | 98.21 | 96.11 | 0.46 | 0.46 | 0.57 | FALCON | 39 |
| 40 | MeilerLabRene | 98.21 | 98.21 | 96.12 | 0.63 | 0.61 | 0.78 | 98.21 | 98.21 | 96.22 | 0.46 | 0.46 | 0.60 | MeilerLabRene | 40 |
| 41 | ZicoFullSTPFullData | 98.21 | 98.21 | 95.47 | 0.63 | 0.61 | 0.68 | 98.21 | 98.21 | 95.47 | 0.46 | 0.46 | 0.45 | ZicoFullSTPFullData | 41 |
| 42 | Pcons_dot_net | 98.21 | 98.21 | 95.48 | 0.63 | 0.61 | 0.68 | 98.21 | 98.21 | 95.54 | 0.46 | 0.46 | 0.46 | Pcons_dot_net | 42 |
| 43 | MUProt | 98.21 | 98.21 | 95.57 | 0.63 | 0.61 | 0.70 | 98.21 | 98.21 | 96.07 | 0.46 | 0.46 | 0.57 | MUProt | 43 |
| 44 | LOOPP_Server | 98.21 | 98.21 | 93.70 | 0.63 | 0.61 | 0.42 | 98.21 | 98.21 | 93.70 | 0.46 | 0.46 | 0.10 | LOOPP_Server | 44 |
| 45 | Phragment | 98.21 | 98.21 | 95.97 | 0.63 | 0.61 | 0.75 | 98.21 | 98.21 | 95.97 | 0.46 | 0.46 | 0.55 | Phragment | 45 |
| 46 | 3DShotMQ | 98.21 | 98.21 | 95.49 | 0.63 | 0.61 | 0.68 | 98.21 | 98.21 | 95.49 | 0.46 | 0.46 | 0.45 | 3DShotMQ | 46 |
| 47 | BioSerf | 98.21 | 98.21 | 93.93 | 0.63 | 0.61 | 0.45 | 98.21 | 98.21 | 93.93 | 0.46 | 0.46 | 0.15 | BioSerf | 47 |
| 48 | KudlatyPredHuman | 98.21 | 98.21 | 95.11 | 0.63 | 0.61 | 0.63 | 98.21 | 98.21 | 95.63 | 0.46 | 0.46 | 0.48 | KudlatyPredHuman | 48 |
| 49 | Jiang_Zhu | 98.21 | 98.21 | 95.68 | 0.63 | 0.61 | 0.71 | 98.21 | 98.21 | 95.68 | 0.46 | 0.46 | 0.49 | Jiang_Zhu | 49 |
| 50 | Chicken_George | 98.21 | 98.21 | 95.17 | 0.63 | 0.61 | 0.64 | 98.21 | 98.21 | 95.17 | 0.46 | 0.46 | 0.39 | Chicken_George | 50 |
| 51 | Bates_BMM | 98.21 | 98.21 | 94.96 | 0.63 | 0.61 | 0.61 | 98.21 | 98.21 | 94.96 | 0.46 | 0.46 | 0.35 | Bates_BMM | 51 |
| 52 | Pcons_multi | 98.21 | 98.21 | 94.12 | 0.63 | 0.61 | 0.48 | 98.21 | 98.21 | 96.21 | 0.46 | 0.46 | 0.59 | Pcons_multi | 52 |
| 53 | RAPTOR | 98.21 | 98.21 | 95.98 | 0.63 | 0.61 | 0.76 | 98.21 | 98.21 | 95.98 | 0.46 | 0.46 | 0.55 | RAPTOR | 53 |
| 54 | MULTICOM | 98.21 | 98.21 | 95.03 | 0.63 | 0.61 | 0.62 | 98.21 | 98.21 | 96.35 | 0.46 | 0.46 | 0.62 | MULTICOM | 54 |
| 55 | MULTICOM−CMFR | 98.21 | 97.02 | 96.20 | 0.63 | 0.47 | 0.79 | 98.21 | 97.02 | 96.20 | 0.46 | 0.26 | 0.59 | MULTICOM−CMFR | 55 |
| 56 | SAMUDRALA | 98.21 | 97.02 | 93.81 | 0.63 | 0.47 | 0.44 | 98.21 | 97.02 | 94.29 | 0.46 | 0.26 | 0.22 | SAMUDRALA | 56 |
| 57 | ZicoFullSTP | 98.21 | 97.02 | 96.22 | 0.63 | 0.47 | 0.79 | 98.21 | 98.21 | 96.22 | 0.46 | 0.46 | 0.60 | ZicoFullSTP | 57 |
| 58 | Frankenstein | 98.21 | 97.02 | 93.49 | 0.63 | 0.47 | 0.39 | 98.21 | 98.21 | 95.40 | 0.46 | 0.46 | 0.44 | Frankenstein | 58 |
| 59 | MULTICOM−REFINE | 98.21 | 97.02 | 96.20 | 0.63 | 0.47 | 0.79 | 98.21 | 98.21 | 96.20 | 0.46 | 0.46 | 0.59 | MULTICOM−REFINE | 59 |
| 60 | pro−sp3−TASSER | 98.21 | 97.02 | 88.59 | 0.63 | 0.47 | 98.21 | 98.21 | 95.07 | 0.46 | 0.46 | 0.37 | pro−sp3−TASSER | 60 | |
| 61 | Zhang−Server | 98.21 | 95.83 | 94.86 | 0.63 | 0.33 | 0.59 | 100.00 | 98.81 | 96.85 | 0.83 | 0.56 | 0.72 | Zhang−Server | 61 |
| 62 | 3DShot2 | 98.21 | 95.83 | 94.33 | 0.63 | 0.33 | 0.51 | 98.21 | 95.83 | 94.33 | 0.46 | 0.06 | 0.23 | 3DShot2 | 62 |
| 63 | PRI−Yang−KiharA | 96.43 | 96.43 | 93.40 | 0.35 | 0.40 | 0.38 | 96.43 | 96.43 | 93.40 | 0.09 | 0.16 | 0.04 | PRI−Yang−KiharA | 63 |
| 64 | PS2−server | 96.43 | 96.43 | 94.39 | 0.35 | 0.40 | 0.52 | 96.43 | 96.43 | 95.07 | 0.09 | 0.16 | 0.37 | PS2−server | 64 |
| 65 | 3DShot1 | 96.43 | 96.43 | 93.93 | 0.35 | 0.40 | 0.45 | 96.43 | 96.43 | 93.93 | 0.09 | 0.16 | 0.15 | 3DShot1 | 65 |
| 66 | fais@hgc | 96.43 | 96.43 | 88.44 | 0.35 | 0.40 | 96.43 | 96.43 | 90.04 | 0.09 | 0.16 | fais@hgc | 66 | ||
| 67 | Bilab−UT | 96.43 | 96.43 | 95.16 | 0.35 | 0.40 | 0.64 | 98.21 | 98.21 | 95.16 | 0.46 | 0.46 | 0.39 | Bilab−UT | 67 |
| 68 | GeneSilico | 96.43 | 96.43 | 92.45 | 0.35 | 0.40 | 0.24 | 98.21 | 98.21 | 95.63 | 0.46 | 0.46 | 0.48 | GeneSilico | 68 |
| 69 | PS2−manual | 96.43 | 96.43 | 94.39 | 0.35 | 0.40 | 0.52 | 96.43 | 96.43 | 95.07 | 0.09 | 0.16 | 0.37 | PS2−manual | 69 |
| 70 | DistillSN | 96.43 | 96.43 | 91.99 | 0.35 | 0.40 | 0.17 | 96.43 | 96.43 | 91.99 | 0.09 | 0.16 | DistillSN | 70 | |
| 71 | A−TASSER | 96.43 | 96.43 | 91.36 | 0.35 | 0.40 | 0.07 | 98.21 | 98.21 | 95.35 | 0.46 | 0.46 | 0.43 | A−TASSER | 71 |
| 72 | MULTICOM−CLUSTER | 96.43 | 95.24 | 94.20 | 0.35 | 0.26 | 0.49 | 96.43 | 96.43 | 94.26 | 0.09 | 0.16 | 0.21 | MULTICOM−CLUSTER | 72 |
| 73 | PSI | 96.43 | 95.24 | 94.88 | 0.35 | 0.26 | 0.59 | 98.21 | 97.02 | 95.98 | 0.46 | 0.26 | 0.55 | PSI | 73 |
| 74 | ACOMPMOD | 96.43 | 95.24 | 94.13 | 0.35 | 0.26 | 0.48 | 96.43 | 95.24 | 94.13 | 0.09 | 0.19 | ACOMPMOD | 74 | |
| 75 | FALCON_CONSENSUS | 94.64 | 94.64 | 92.44 | 0.08 | 0.19 | 0.23 | 96.43 | 96.43 | 92.67 | 0.09 | 0.16 | FALCON_CONSENSUS | 75 | |
| 76 | TJ_Jiang | 94.64 | 94.64 | 90.68 | 0.08 | 0.19 | 98.21 | 98.21 | 95.37 | 0.46 | 0.46 | 0.43 | TJ_Jiang | 76 | |
| 77 | MidwayFolding | 94.64 | 94.64 | 92.82 | 0.08 | 0.19 | 0.29 | 98.21 | 98.21 | 95.97 | 0.46 | 0.46 | 0.55 | MidwayFolding | 77 |
| 78 | igor | 94.64 | 94.64 | 91.78 | 0.08 | 0.19 | 0.14 | 94.64 | 94.64 | 91.78 | igor | 78 | |||
| 79 | POEM | 94.64 | 93.45 | 91.64 | 0.08 | 0.05 | 0.12 | 98.21 | 98.21 | 96.22 | 0.46 | 0.46 | 0.60 | POEM | 79 |
| 80 | JIVE08 | 92.86 | 92.86 | 90.08 | 98.21 | 98.21 | 93.27 | 0.46 | 0.46 | 0.02 | JIVE08 | 80 | |||
| 81 | Kolinski | 92.86 | 92.86 | 93.12 | 0.33 | 98.21 | 98.21 | 95.07 | 0.46 | 0.46 | 0.37 | Kolinski | 81 | ||
| 82 | fais−server | 92.86 | 92.86 | 87.48 | 96.43 | 96.43 | 93.98 | 0.09 | 0.16 | 0.16 | fais−server | 82 | |||
| 83 | pipe_int | 92.86 | 92.86 | 87.96 | 92.86 | 92.86 | 87.96 | pipe_int | 83 | ||||||
| 84 | EB_AMU_Physics | 92.86 | 92.86 | 90.73 | 92.86 | 92.86 | 90.73 | EB_AMU_Physics | 84 | ||||||
| 85 | Sternberg | 92.86 | 91.67 | 87.14 | 98.21 | 98.21 | 95.97 | 0.46 | 0.46 | 0.55 | Sternberg | 85 | |||
| 86 | Poing | 92.86 | 91.67 | 87.14 | 92.86 | 91.67 | 91.23 | Poing | 86 | ||||||
| 87 | Phyre_de_novo | 92.86 | 91.67 | 87.14 | 92.86 | 91.67 | 91.23 | Phyre_de_novo | 87 | ||||||
| 88 | Pcons_local | 92.86 | 90.48 | 92.30 | 0.21 | 98.21 | 98.21 | 94.95 | 0.46 | 0.46 | 0.35 | Pcons_local | 88 | ||
| 89 | Hao_Kihara | 91.07 | 91.07 | 89.50 | 92.86 | 92.86 | 93.33 | 0.03 | Hao_Kihara | 89 | |||||
| 90 | ABIpro | 91.07 | 91.07 | 91.14 | 0.04 | 98.21 | 98.21 | 94.42 | 0.46 | 0.46 | 0.24 | ABIpro | 90 | ||
| 91 | FUGUE_KM | 91.07 | 89.88 | 89.18 | 98.21 | 98.21 | 95.47 | 0.46 | 0.46 | 0.45 | FUGUE_KM | 91 | |||
| 92 | mGenTHREADER | 91.07 | 88.69 | 89.21 | 91.07 | 88.69 | 89.21 | mGenTHREADER | 92 | ||||||
| 93 | nFOLD3 | 89.29 | 89.29 | 88.88 | 98.21 | 98.21 | 96.70 | 0.46 | 0.46 | 0.69 | nFOLD3 | 93 | |||
| 94 | MUFOLD−MD | 89.29 | 89.29 | 86.38 | 96.43 | 96.43 | 93.34 | 0.09 | 0.16 | 0.03 | MUFOLD−MD | 94 | |||
| 95 | Handl−Lovell | 89.29 | 89.29 | 89.11 | 98.21 | 98.21 | 96.00 | 0.46 | 0.46 | 0.55 | Handl−Lovell | 95 | |||
| 96 | 3Dpro | 87.50 | 82.74 | 86.03 | 89.29 | 86.91 | 86.63 | 3Dpro | 96 | ||||||
| 97 | POISE | 87.50 | 80.36 | 85.82 | 92.86 | 89.29 | 90.53 | POISE | 97 | ||||||
| 98 | Wolynes | 85.71 | 82.14 | 83.41 | 96.43 | 96.43 | 94.03 | 0.09 | 0.16 | 0.17 | Wolynes | 98 | |||
| 99 | ShakAbInitio | 83.93 | 83.93 | 73.16 | 83.93 | 83.93 | 80.30 | ShakAbInitio | 99 | ||||||
| 100 | Pushchino | 83.93 | 83.93 | 85.31 | 83.93 | 83.93 | 85.31 | Pushchino | 100 | ||||||
| 101 | FEIG | 83.93 | 82.74 | 80.84 | 98.21 | 97.02 | 94.56 | 0.46 | 0.26 | 0.27 | FEIG | 101 | |||
| 102 | FAMSD | 83.93 | 77.98 | 81.00 | 89.29 | 77.98 | 84.98 | FAMSD | 102 | ||||||
| 103 | circle | 83.93 | 76.79 | 82.04 | 98.21 | 98.21 | 94.25 | 0.46 | 0.46 | 0.21 | circle | 103 | |||
| 104 | 3D−JIGSAW_AEP | 82.14 | 82.14 | 75.50 | 82.14 | 82.14 | 75.50 | 3D−JIGSAW_AEP | 104 | ||||||
| 105 | CpHModels | 82.14 | 80.95 | 76.93 | 82.14 | 80.95 | 76.93 | CpHModels | 105 | ||||||
| 106 | RPFM | 82.14 | 77.38 | 70.95 | 83.93 | 82.74 | 86.51 | RPFM | 106 | ||||||
| 107 | 3D−JIGSAW_V3 | 80.36 | 79.17 | 76.70 | 82.14 | 80.95 | 76.70 | 3D−JIGSAW_V3 | 107 | ||||||
| 108 | schenk−torda−server | 80.36 | 63.69 | 75.53 | 89.29 | 88.09 | 82.38 | schenk−torda−server | 108 | ||||||
| 109 | mahmood−torda−server | 80.36 | 61.31 | 75.85 | 80.36 | 61.91 | 75.85 | mahmood−torda−server | 109 | ||||||
| 110 | mariner1 | 78.57 | 64.29 | 78.93 | 87.50 | 87.50 | 91.37 | mariner1 | 110 | ||||||
| 111 | Softberry | 75.00 | 69.05 | 68.28 | 75.00 | 69.05 | 68.28 | Softberry | 111 | ||||||
| 112 | COMA−M | 75.00 | 67.86 | 69.47 | 78.57 | 73.21 | 74.77 | COMA−M | 112 | ||||||
| 113 | IBT_LT | 73.21 | 69.64 | 69.74 | 73.21 | 69.64 | 69.74 | IBT_LT | 113 | ||||||
| 114 | COMA | 73.21 | 67.26 | 69.11 | 80.36 | 74.41 | 74.77 | COMA | 114 | ||||||
| 115 | RANDOM | 70.61 | 68.80 | 70.61 | 70.61 | 68.80 | 70.61 | RANDOM | 115 | ||||||
| 116 | DelCLab | 69.64 | 68.45 | 69.60 | 69.64 | 68.45 | 69.65 | DelCLab | 116 | ||||||
| 117 | StruPPi | 67.86 | 54.76 | 71.52 | 67.86 | 54.76 | 71.52 | StruPPi | 117 | ||||||
| 118 | HHpred5 | 64.29 | 64.29 | 67.52 | 64.29 | 64.29 | 67.52 | HHpred5 | 118 | ||||||
| 119 | FOLDpro | 64.29 | 55.95 | 43.03 | 89.29 | 86.91 | 86.03 | FOLDpro | 119 | ||||||
| 120 | SMEG−CCP | 64.29 | 53.57 | 75.21 | 64.29 | 53.57 | 75.21 | SMEG−CCP | 120 | ||||||
| 121 | HHpred4 | 64.29 | 50.00 | 63.11 | 64.29 | 50.00 | 63.11 | HHpred4 | 121 | ||||||
| 122 | HHpred2 | 60.71 | 48.81 | 43.48 | 60.71 | 48.81 | 43.48 | HHpred2 | 122 | ||||||
| 123 | TWPPLAB | 58.93 | 57.74 | 41.62 | 58.93 | 57.74 | 41.62 | TWPPLAB | 123 | ||||||
| 124 | rehtnap | 39.29 | 32.14 | 29.38 | 39.29 | 32.14 | 29.38 | rehtnap | 124 | ||||||
| 125 | FFASstandard | 35.71 | 35.71 | 32.15 | 35.71 | 35.71 | 32.15 | FFASstandard | 125 | ||||||
| 126 | FFASflextemplate | 35.71 | 35.71 | 32.15 | 35.71 | 35.71 | 32.15 | FFASflextemplate | 126 | ||||||
| 127 | FFASsuboptimal | 35.71 | 35.71 | 32.04 | 35.71 | 35.71 | 32.30 | FFASsuboptimal | 127 | ||||||
| 128 | SAM−T02−server | 0.00 | 0.00 | 19.38 | 98.21 | 98.21 | 95.51 | 0.46 | 0.46 | 0.46 | SAM−T02−server | 128 | |||
| 129 | Abagyan | Abagyan | 129 | ||||||||||||
| 130 | BHAGEERATH | BHAGEERATH | 130 | ||||||||||||
| 131 | CBSU | CBSU | 131 | ||||||||||||
| 132 | FEIG_REFINE | FEIG_REFINE | 132 | ||||||||||||
| 133 | Fiser−M4T | Fiser−M4T | 133 | ||||||||||||
| 134 | FrankensteinLong | FrankensteinLong | 134 | ||||||||||||
| 135 | HCA | HCA | 135 | ||||||||||||
| 136 | LEE−SERVER | LEE−SERVER | 136 | ||||||||||||
| 137 | Linnolt−UH−CMB | Linnolt−UH−CMB | 137 | ||||||||||||
| 138 | NIM2 | NIM2 | 138 | ||||||||||||
| 139 | Nano_team | Nano_team | 139 | ||||||||||||
| 140 | NirBenTal | NirBenTal | 140 | ||||||||||||
| 141 | PHAISTOS | PHAISTOS | 141 | ||||||||||||
| 142 | PZ−UAM | PZ−UAM | 142 | ||||||||||||
| 143 | ProtAnG | ProtAnG | 143 | ||||||||||||
| 144 | ProteinShop | ProteinShop | 144 | ||||||||||||
| 145 | Scheraga | Scheraga | 145 | ||||||||||||
| 146 | TsaiLab | TsaiLab | 146 | ||||||||||||
| 147 | UCDavisGenome | UCDavisGenome | 147 | ||||||||||||
| 148 | Wolfson−FOBIA | Wolfson−FOBIA | 148 | ||||||||||||
| 149 | YASARA | YASARA | 149 | ||||||||||||
| 150 | YASARARefine | YASARARefine | 150 | ||||||||||||
| 151 | Zhou−SPARKS | Zhou−SPARKS | 151 | ||||||||||||
| 152 | dill_ucsf | dill_ucsf | 152 | ||||||||||||
| 153 | dill_ucsf_extended | dill_ucsf_extended | 153 | ||||||||||||
| 154 | forecast | forecast | 154 | ||||||||||||
| 155 | huber−torda−server | huber−torda−server | 155 | ||||||||||||
| 156 | jacobson | jacobson | 156 | ||||||||||||
| 157 | mti | mti | 157 | ||||||||||||
| 158 | mumssp | mumssp | 158 | ||||||||||||
| 159 | panther_server | panther_server | 159 | ||||||||||||
| 160 | psiphifoldings | psiphifoldings | 160 | ||||||||||||
| 161 | ricardo | ricardo | 161 | ||||||||||||
| 162 | rivilo | rivilo | 162 | ||||||||||||
| 163 | sessions | sessions | 163 | ||||||||||||
| 164 | taylor | taylor | 164 | ||||||||||||
| 165 | test_http_server_01 | test_http_server_01 | 165 | ||||||||||||
| 166 | tripos_08 | tripos_08 | 166 | ||||||||||||