T0473
protein A3DK08 from Clostridium thermocellum
Target sequence:
>T0473 CmR9, Clostridium thermocellum, 68 residues
MKITKDMIIADVLQMDRGTAPIFINNGMHCLGCPSSMGESIEDACAVHGIDADKLVKELNEYFEKKEV
Structure:
Method: NMR
Determined by: NESG
PDB ID: 2k53
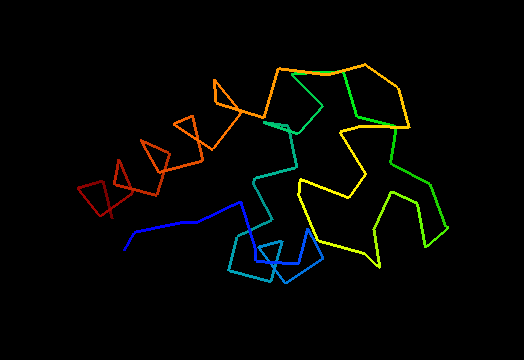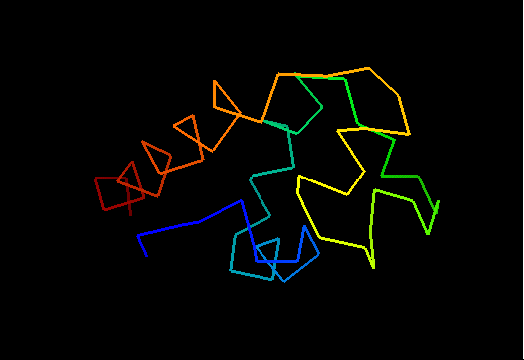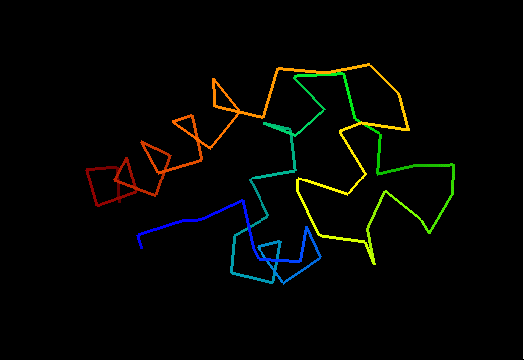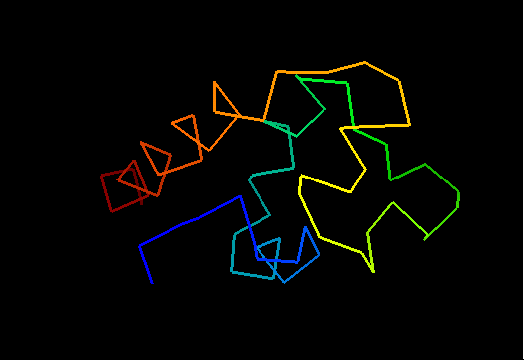
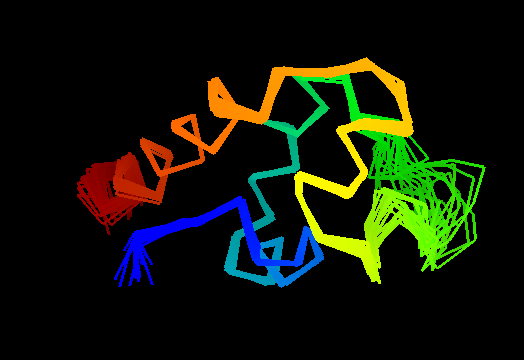
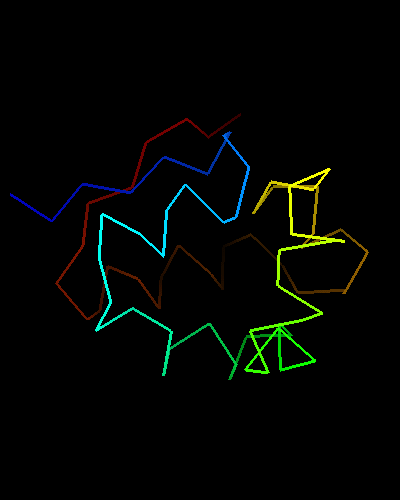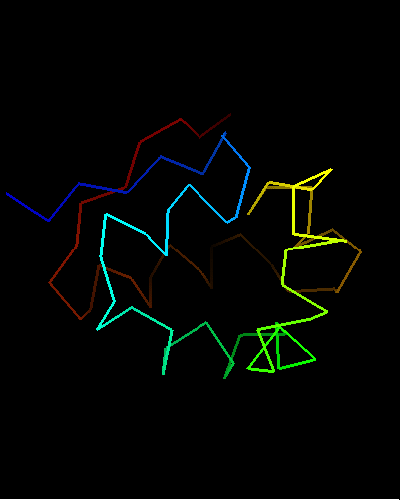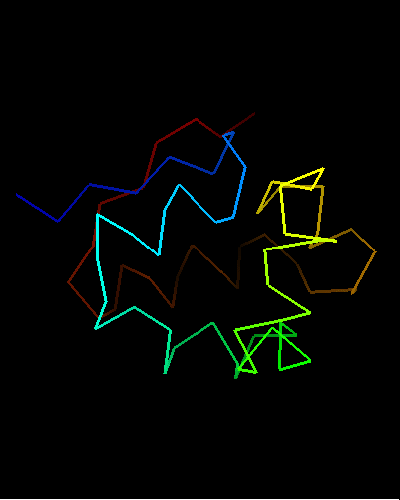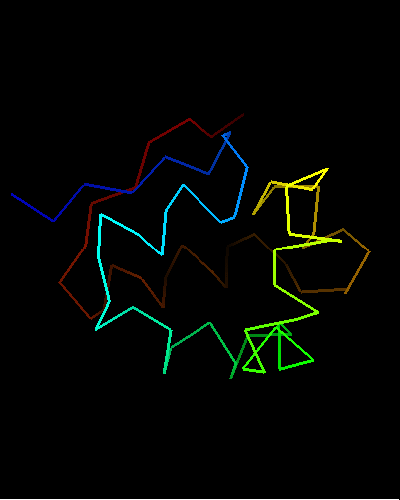
Domains: PyMOL of domains
Single domain protein. Unfortunately, the most interesting region (differing from the template) is conformationally flexible in the middle (green in the image above) of this NMR structure, and some residues from it were removed from NMR models for evaluation of predictions.
Structure classification:
An unusual helix-turn-helix (HTH) containing protein. The last two α-helices (yellow and orange-red) form the HTH motif, which is readily recognizable by almost perpendicular orientation of the two helices and a long extended "turn" between them that positions the N-terminus of the second helix close to the middle of the first helix. The rest of the structure is unique and shared only with the closest homologs: T0469 and the only full-chain template 2fi0. This N-terminal segment is structured as 3 helices and completes the HTH core in a manner similar to how a single N-terminal helix packs agaist HTH in a classic three-helical bundle fold.
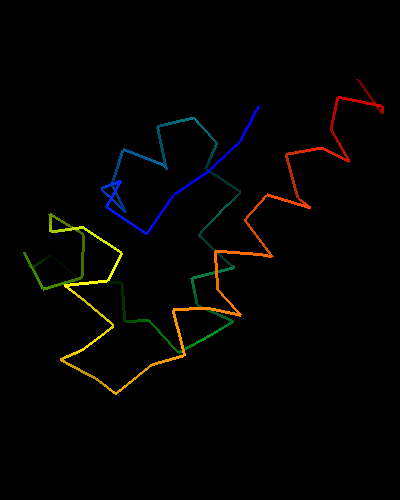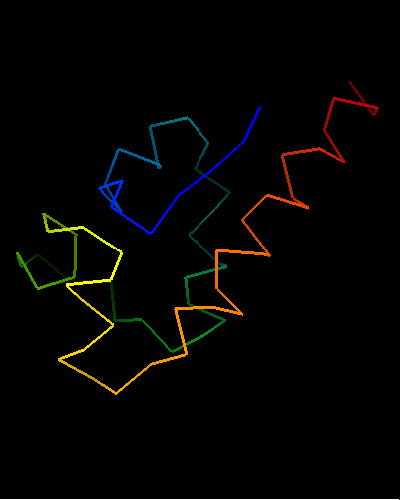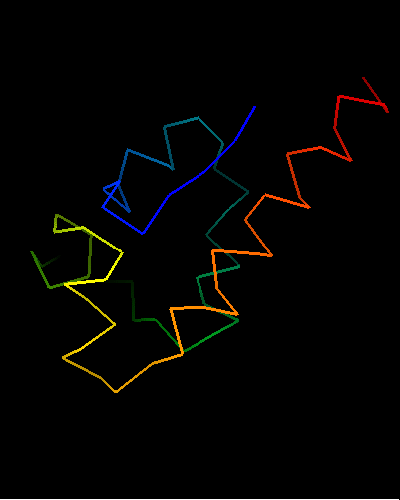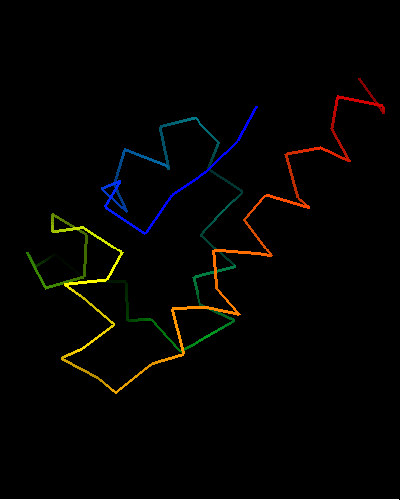
Ribbon diagram of T0473: 2k53 model 1
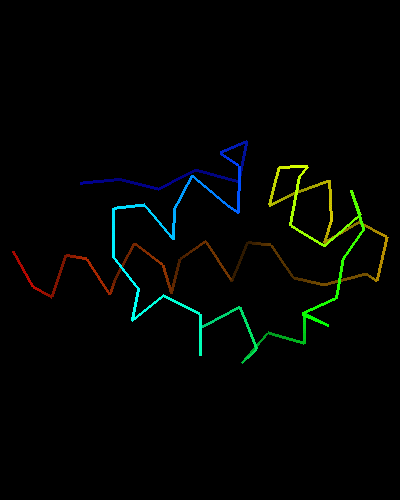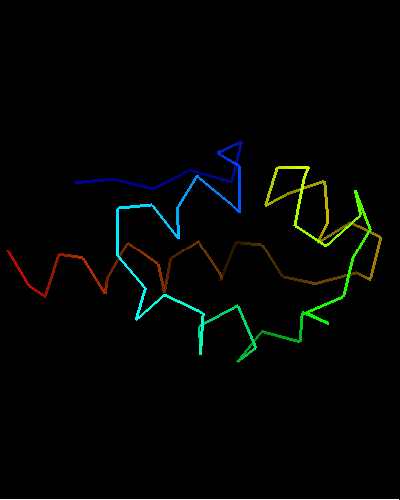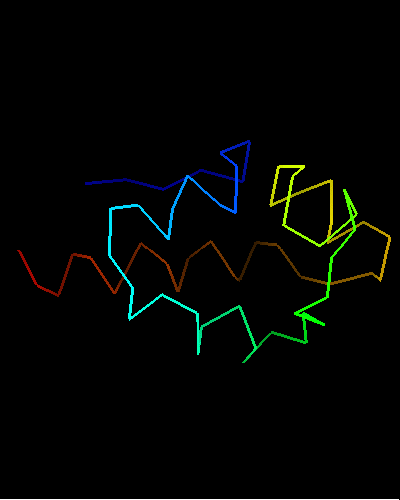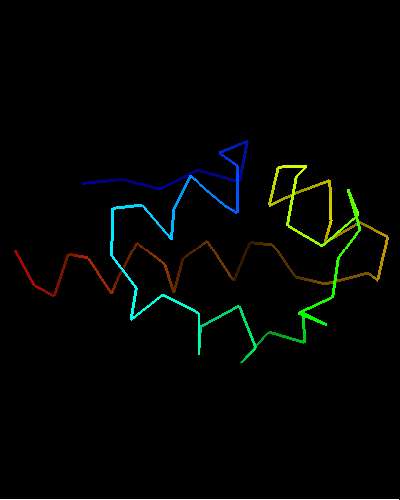
View of T0473 from the opposite side, residues 1 to 67
We used COMPASS to find homologs of T0473. The first COMPASS hit in the SCOP database is to a typical HTH three-helical bundle. Reasonably small E-value of ~0.03 is almost good enough to validate this match.
Subject = d1hcra_ a.4.1.2 HIN recombinase (DNA-binding domain) length=52 filtered_length=51 Neff=12.765 Smith-Waterman score = 48 Evalue = 2.80e-02 QUERY 21 PIFINNGMHCLGCPSSMGESIEDACAVHGIDADKLVKELNEYFEKKE 67 CONSENSUS_1 21 EVFLKYGLDCCGCPIAGFETLEDACREHGIDLDELLEELNEAIAEKD 67 +++ + ++ + +A+ E++++++RE G+ ++ L + L++ ++++ CONSENSUS_2 4 PKLTPEQIAEARRLLAAGESVAEIARELGVSRSTLYRYLPASEKRTA 50 d1hcra_ 4 RAINKHEQEQISRLLEKGHPRQQLAIIFGIGVSTLYRYFPASSIKKR 50
All top COMPASS hits are HTH-containing proteins with different SCOP folds. Additionally, their E-values are quite low, so there is little doubt about evolutionary origin of T0473 and its homology to HTH domains. The 49-Gly-Ile-Asp-51 in T0473 is a very typical sequence for an HTH turn, and Ala44, four residues before Gly49, is a conserved small residue to make room for the second HTH helix packing. However only the HTH part of the domain is homologous, as the way it is completed is quite different among homologs:

Ribbon diagram of T0473: 2k53 model 1
HTH is yellow-orange-red
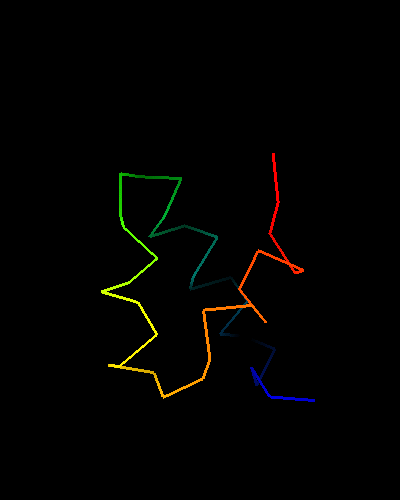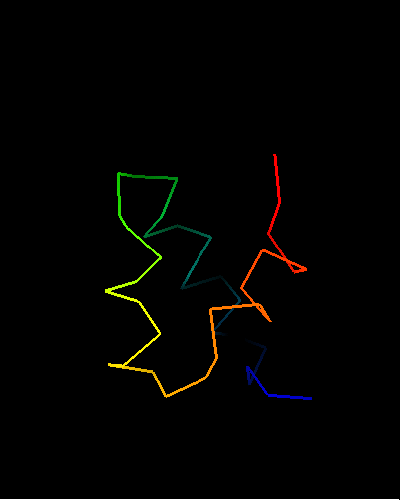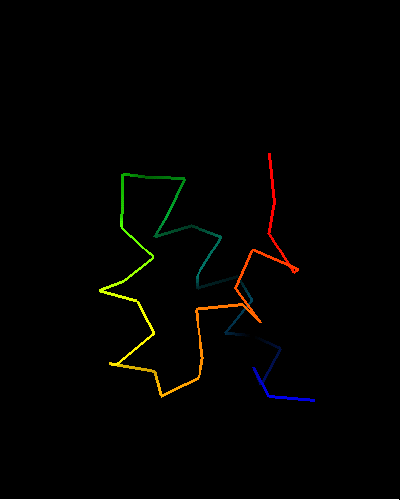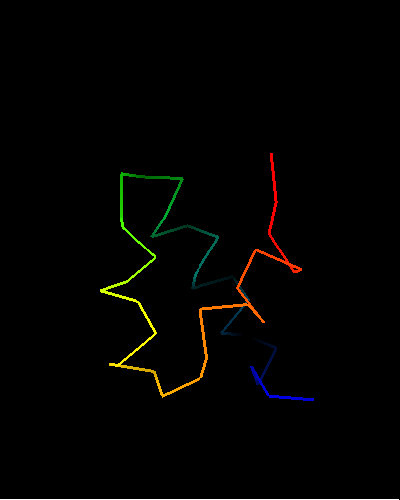
HIN recombinase DNA-binding domain: 1hcr chain A residues 142-182;
HTH is lime-yellow-orange-red
CASP category:
Comparative modeling: medium. Closest template detectable by HHpred, but contains some structural differences.
Closest templates:
The only full-length template was 2fi0, which is another structure of a hypothetical protein determined by a structural genomics center.
T0473 ----MKITKDMIIADVLQMDRGTAPIFINNGMHCLGCP-----SSMGESIEDACAVHGIDADKLVKELNEYFEKKEV--
2fi0_A VVMDNIIDVSIPVAEVVDKHPEVLEILVELGFKPLANPLMRNTVGRKVSLKQGSKLAGTPMDKIVRTLEANGYEVIGLD
Both molecules are shown below with the most similar HTH region (yellow-red) at the back and more variable N-terminal helices in front. The first two helices (blue-cyan and cyan-green) have very similar orientations in both molecules, however, the differences are pronounced in the green-yellow region, where T0473 has a 6-residue deletion eliminating the fourth helix of 2fi0. Another interesting difference is that 2fi0 contains a C-terminal segment (red) that forms β-strand pairing with the N-terminial segment (blue). Thus a two-stranded β-sheet is present in 2fi0. However, T0473 lacks the C-terminal extension. Nevertheless, the N-terminal region (blue) still holds the extended conformation even without H-bond pairing, at least close to the first helix.
Target sequence - PDB file inconsistencies:
NMR models contain two C-terminal residues 69-LE-70, not present in the target sequence (coming from His-tag).
T0473 2k53.pdb T0473.pdb PyMOL PyMOL of domains
T0473 1 MKITKDMIIADVLQMDRGTAPIFINNGMHCLGCPSSMGESIEDACAVHGIDADKLVKELNEYFEKKEV-- 68 ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~ 2k53A 1 MKITKDMIIADVLQMDRGTAPIFINNGMHCLGCPSSMGESIEDACAVHGIDADKLVKELNEYFEKKEVLE 70
Single domain protein: target 1-68 ; pdb 1-68
Sequence classification:
COG2846 in COG database, which is predicted to be "Regulator of cell morphogenesis and NO signaling". Below is PROMALS3D alignment of several most distant homologs detectable by PSI-BLAST. This alignment was not edited manually and may have errors.
Conservation: 9 5 7 6 5 7 T0473 ----MKITKDMIIADVLQMDRGTAPIFINNGMHCLGCP-----SSMGESIEDACAVHGIDADKLVKELNEYFEKKEV-------------------------------------
T0469 1 --MTQKFTKDMTFAQALQTHPGVAGVLRSYNLGCIGCM-----GAQNESLEQGANAHGLNVEDILRDLNALA------------------------------------------ 65
2fi0_chainA 1 VVMDNIIDVSIPVAEVVDKHPEVLEILVELGFKPLANPLMRNTVGRKVSLKQGSKLAGTPMDKIVRTLEANGYEVIGLD----------------------------------- 79
gi_81428069 1 --MAKQIKLSQTVYELVTAYPEIVPILVSIGLDGVTNPALLNTAGRFMTLPKGAQMKQIPLSQIVATLTENDFEVIADD----------------------------------- 77
gi_109645979 1 --MA-LITKDMTVGQVLRSYPQTVQTFLELGMHCLGCPS-----STMESIEGAALTHGKKPDELVEKLNKVI----AAN----------------------------------- 67
gi_95930439 1 ----MLIQPQMTIKQIIDTWPQTLDVFTANGFEQFAQPEMIDRVGRFLKLESALSQKGYDCAAFIALLQERI----DAADQQADI-------TLVE------------------ 81
gi_146300169 1 --M--ENLKNKTIGSFVAQDYRTAAVFSKYRIDF-CCK-------GNRTVDEVCEKQNIDADVLLQNIHEVI----QSENNGSIDFNSWPLDLLADYIEKTHHRYVEDKTHTLL 98
gi_163939963 1 --MKHTFTETSIIGEIVTQFPKASDLFKSYRIDF-CCG-------GNRPLIDAINERNLSATEVITELNTLY-NNTKLLNESEIDWKNASYRELIDYVINKHHR---------- 93
Consensus_aa: ..M..bhp.sbhl.phlpp@Pphh.lh.ph.hc..ts...........sl.pthp.pshshs.ll.pLp............................................. Consensus_ss: hhhhhhh hhhhhhh hhhhhhhh hhhhhhhhhhhh
Comments:
This protein is a close homolog of T0469:
T0473 --MKITKDMIIADVLQMDRGTAPIFINNGMHCLGCPSSMGESIEDACAVHGIDADKLVKELNEYFEKKEVT0469 MTQKFTKDMTFAQALQTHPGVAGVLRSYNLGCIGCMGAQNESLEQGANAHGLNVEDILRDLNALA-----
Curiously, many sequences of this family contain conserved Cysteine residues in the most varible region of the molecule, and some also contain Histidine in the HTH turn. These residues are highlighted in magenta in the above family alignment and are shown on the structure of T0473. Due to their conservation, it was hypothesized that they might be part of a metal or some other ligand - binding site.
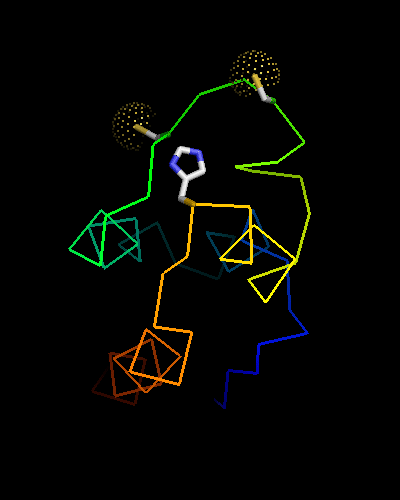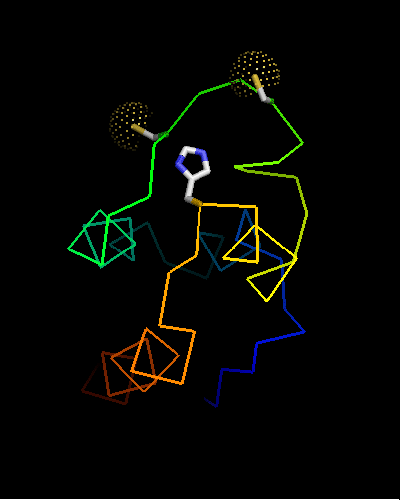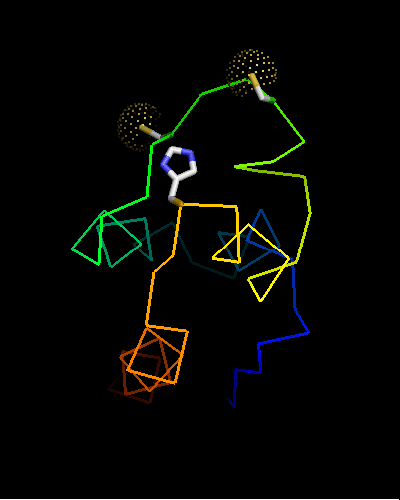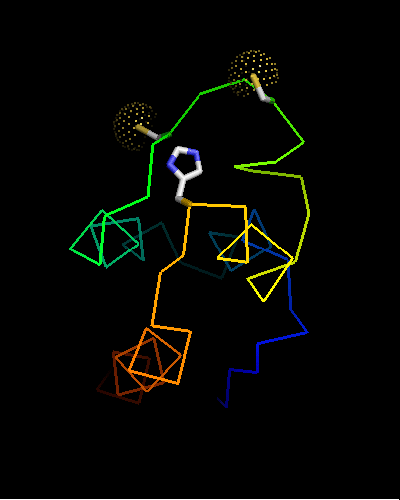
Ribbon diagram of T0473: 2k53 model 15,
Cys30, Cys33 and His48 are shown
Server predictions:
T0473:pdb 1-68:seq 1-68:CM_medium; alignment
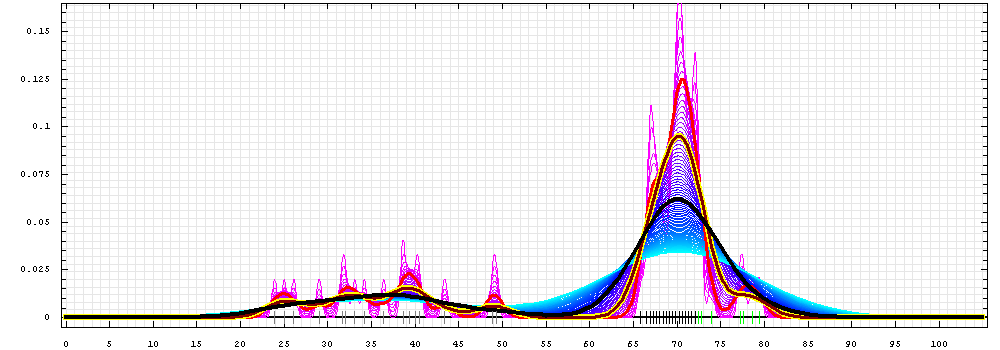
First models for T0473:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0473 | T0473 | T0473 | T0473 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | mufold | 81.25 | 77.33 | 80.82 | 1.32 | 1.73 | 1.57 | 82.72 | 79.78 | 80.82 | 1.42 | 1.89 | 1.41 | mufold | 1 |
| 2 | KudlatyPredHuman | 79.41 | 70.59 | 79.18 | 1.17 | 1.16 | 1.39 | 79.41 | 72.18 | 80.68 | 1.11 | 1.18 | 1.39 | KudlatyPredHuman | 2 |
| 3 | METATASSER | 79.41 | 69.61 | 77.58 | 1.17 | 1.08 | 1.21 | 79.41 | 71.32 | 77.58 | 1.11 | 1.09 | 1.03 | METATASSER | 3 |
| 4 | MULTICOM | 78.68 | 72.06 | 79.38 | 1.11 | 1.29 | 1.41 | 78.68 | 72.06 | 79.70 | 1.04 | 1.16 | 1.28 | MULTICOM | 4 |
| 5 | DBAKER | 78.68 | 69.36 | 80.46 | 1.11 | 1.06 | 1.53 | 78.68 | 69.36 | 80.46 | 1.04 | 0.91 | 1.37 | DBAKER | 5 |
| 6 | Zhang−Server | 77.57 | 69.73 | 78.26 | 1.02 | 1.09 | 1.28 | 79.04 | 71.94 | 78.81 | 1.08 | 1.15 | 1.17 | Zhang−Server | 6 |
| 7 | GeneSilico | 77.57 | 69.73 | 78.26 | 1.02 | 1.09 | 1.28 | 77.57 | 69.73 | 78.26 | 0.94 | 0.94 | 1.11 | GeneSilico | 7 |
| 8 | McGuffin | 77.57 | 69.73 | 78.26 | 1.02 | 1.09 | 1.28 | 82.35 | 79.41 | 78.60 | 1.39 | 1.86 | 1.15 | McGuffin | 8 |
| 9 | FEIG | 77.21 | 70.59 | 69.16 | 0.99 | 1.16 | 0.26 | 77.21 | 70.59 | 72.28 | 0.91 | 1.03 | 0.40 | FEIG | 9 |
| 10 | Zhang | 76.84 | 71.69 | 78.02 | 0.96 | 1.25 | 1.26 | 78.31 | 72.55 | 78.55 | 1.01 | 1.21 | 1.14 | Zhang | 10 |
| 11 | TASSER | 75.73 | 67.65 | 77.76 | 0.87 | 0.91 | 1.23 | 75.73 | 67.65 | 77.76 | 0.77 | 0.75 | 1.05 | TASSER | 11 |
| 12 | AMU−Biology | 74.63 | 70.47 | 69.44 | 0.78 | 1.15 | 0.29 | 75.37 | 70.47 | 74.49 | 0.74 | 1.01 | 0.66 | AMU−Biology | 12 |
| 13 | Sternberg | 73.90 | 66.54 | 74.01 | 0.72 | 0.82 | 0.80 | 73.90 | 66.54 | 74.01 | 0.60 | 0.64 | 0.60 | Sternberg | 13 |
| 14 | Phyre_de_novo | 73.90 | 66.54 | 74.02 | 0.72 | 0.82 | 0.81 | 73.90 | 66.54 | 74.05 | 0.60 | 0.64 | 0.61 | Phyre_de_novo | 14 |
| 15 | SAM−T08−human | 73.90 | 63.85 | 77.74 | 0.72 | 0.59 | 1.23 | 73.90 | 66.91 | 79.06 | 0.60 | 0.68 | 1.20 | SAM−T08−human | 15 |
| 16 | StruPPi | 73.16 | 65.81 | 69.98 | 0.66 | 0.75 | 0.35 | 73.16 | 65.81 | 69.98 | 0.53 | 0.57 | 0.12 | StruPPi | 16 |
| 17 | 3DShotMQ | 73.16 | 65.56 | 75.94 | 0.66 | 0.73 | 1.02 | 73.16 | 65.56 | 75.94 | 0.53 | 0.55 | 0.83 | 3DShotMQ | 17 |
| 18 | Bates_BMM | 73.16 | 59.19 | 76.31 | 0.66 | 0.19 | 1.06 | 79.41 | 74.27 | 79.86 | 1.11 | 1.37 | 1.30 | Bates_BMM | 18 |
| 19 | 3D−JIGSAW_V3 | 72.79 | 62.99 | 77.32 | 0.63 | 0.51 | 1.18 | 73.90 | 68.63 | 77.32 | 0.60 | 0.84 | 0.99 | 3D−JIGSAW_V3 | 19 |
| 20 | IBT_LT | 72.79 | 62.26 | 73.88 | 0.63 | 0.45 | 0.79 | 72.79 | 62.26 | 73.88 | 0.49 | 0.24 | 0.59 | IBT_LT | 20 |
| 21 | LevittGroup | 72.43 | 68.02 | 69.39 | 0.60 | 0.94 | 0.28 | 73.53 | 68.02 | 72.04 | 0.56 | 0.78 | 0.37 | LevittGroup | 21 |
| 22 | BAKER−ROBETTA | 72.43 | 60.91 | 72.27 | 0.60 | 0.34 | 0.61 | 74.27 | 65.93 | 75.62 | 0.63 | 0.58 | 0.79 | BAKER−ROBETTA | 22 |
| 23 | GS−KudlatyPred | 72.43 | 60.91 | 72.27 | 0.60 | 0.34 | 0.61 | 72.43 | 60.91 | 72.27 | 0.46 | 0.11 | 0.39 | GS−KudlatyPred | 23 |
| 24 | Poing | 72.06 | 65.44 | 71.92 | 0.57 | 0.72 | 0.57 | 72.06 | 65.44 | 71.92 | 0.43 | 0.54 | 0.35 | Poing | 24 |
| 25 | FAMS−multi | 72.06 | 64.71 | 71.42 | 0.57 | 0.66 | 0.51 | 72.43 | 64.71 | 71.66 | 0.46 | 0.47 | 0.32 | FAMS−multi | 25 |
| 26 | MUSTER | 72.06 | 64.71 | 69.36 | 0.57 | 0.66 | 0.28 | 72.06 | 64.71 | 69.36 | 0.43 | 0.47 | 0.05 | MUSTER | 26 |
| 27 | SAM−T08−server | 72.06 | 64.46 | 77.93 | 0.57 | 0.64 | 1.25 | 74.27 | 66.42 | 77.93 | 0.63 | 0.63 | 1.07 | SAM−T08−server | 27 |
| 28 | BioSerf | 72.06 | 58.82 | 70.80 | 0.57 | 0.16 | 0.44 | 72.06 | 58.82 | 70.80 | 0.43 | 0.22 | BioSerf | 28 | |
| 29 | ACOMPMOD | 72.06 | 56.37 | 72.56 | 0.57 | 0.64 | 72.06 | 56.37 | 72.56 | 0.43 | 0.43 | ACOMPMOD | 29 | ||
| 30 | LEE | 72.06 | 53.68 | 75.59 | 0.57 | 0.98 | 72.79 | 62.38 | 75.59 | 0.49 | 0.25 | 0.79 | LEE | 30 | |
| 31 | 3D−JIGSAW_AEP | 71.69 | 68.50 | 70.36 | 0.54 | 0.98 | 0.39 | 73.16 | 69.00 | 72.54 | 0.53 | 0.87 | 0.43 | 3D−JIGSAW_AEP | 31 |
| 32 | NirBenTal | 71.69 | 65.32 | 70.06 | 0.54 | 0.71 | 0.36 | 71.69 | 65.32 | 70.06 | 0.39 | 0.53 | 0.13 | NirBenTal | 32 |
| 33 | fams−ace2 | 71.69 | 64.09 | 77.66 | 0.54 | 0.61 | 1.22 | 71.69 | 64.09 | 77.66 | 0.39 | 0.41 | 1.04 | fams−ace2 | 33 |
| 34 | pro−sp3−TASSER | 71.69 | 63.85 | 72.92 | 0.54 | 0.59 | 0.68 | 82.35 | 79.41 | 77.66 | 1.39 | 1.86 | 1.04 | pro−sp3−TASSER | 34 |
| 35 | EB_AMU_Physics | 71.69 | 62.62 | 71.97 | 0.54 | 0.48 | 0.57 | 71.69 | 62.62 | 71.97 | 0.39 | 0.27 | 0.36 | EB_AMU_Physics | 35 |
| 36 | tripos_08 | 71.32 | 65.93 | 65.42 | 0.51 | 0.76 | 71.32 | 65.93 | 65.42 | 0.36 | 0.58 | tripos_08 | 36 | ||
| 37 | Phragment | 71.32 | 65.20 | 71.09 | 0.51 | 0.70 | 0.47 | 71.32 | 65.20 | 71.09 | 0.36 | 0.52 | 0.25 | Phragment | 37 |
| 38 | Zico | 71.32 | 64.71 | 72.80 | 0.51 | 0.66 | 0.67 | 77.57 | 69.48 | 78.68 | 0.94 | 0.92 | 1.16 | Zico | 38 |
| 39 | ZicoFullSTP | 71.32 | 64.71 | 72.80 | 0.51 | 0.66 | 0.67 | 77.57 | 69.48 | 78.68 | 0.94 | 0.92 | 1.16 | ZicoFullSTP | 39 |
| 40 | ZicoFullSTPFullData | 71.32 | 64.71 | 72.80 | 0.51 | 0.66 | 0.67 | 72.79 | 64.71 | 78.68 | 0.49 | 0.47 | 1.16 | ZicoFullSTPFullData | 40 |
| 41 | keasar−server | 71.32 | 62.74 | 70.65 | 0.51 | 0.49 | 0.42 | 72.43 | 63.85 | 72.36 | 0.46 | 0.39 | 0.41 | keasar−server | 41 |
| 42 | Phyre2 | 70.96 | 63.36 | 71.61 | 0.48 | 0.55 | 0.53 | 70.96 | 63.36 | 71.61 | 0.32 | 0.34 | 0.32 | Phyre2 | 42 |
| 43 | Hao_Kihara | 70.96 | 63.11 | 69.42 | 0.48 | 0.53 | 0.29 | 70.96 | 63.11 | 69.42 | 0.32 | 0.32 | 0.06 | Hao_Kihara | 43 |
| 44 | MidwayFolding | 70.96 | 63.11 | 71.78 | 0.48 | 0.53 | 0.55 | 70.96 | 63.11 | 71.78 | 0.32 | 0.32 | 0.34 | MidwayFolding | 44 |
| 45 | RAPTOR | 70.96 | 63.11 | 71.78 | 0.48 | 0.53 | 0.55 | 71.69 | 63.11 | 72.68 | 0.39 | 0.32 | 0.44 | RAPTOR | 45 |
| 46 | PS2−server | 70.96 | 62.38 | 70.21 | 0.48 | 0.46 | 0.37 | 70.96 | 62.38 | 70.21 | 0.32 | 0.25 | 0.15 | PS2−server | 46 |
| 47 | PS2−manual | 70.96 | 62.38 | 70.21 | 0.48 | 0.46 | 0.37 | 70.96 | 62.38 | 70.21 | 0.32 | 0.25 | 0.15 | PS2−manual | 47 |
| 48 | Bilab−UT | 70.96 | 60.66 | 71.84 | 0.48 | 0.32 | 0.56 | 72.43 | 62.74 | 71.84 | 0.46 | 0.28 | 0.34 | Bilab−UT | 48 |
| 49 | Kolinski | 70.59 | 62.01 | 74.00 | 0.45 | 0.43 | 0.80 | 70.96 | 63.36 | 75.08 | 0.32 | 0.34 | 0.73 | Kolinski | 49 |
| 50 | COMA−M | 70.59 | 61.52 | 68.61 | 0.45 | 0.39 | 0.19 | 70.59 | 61.52 | 68.61 | 0.29 | 0.17 | COMA−M | 50 | |
| 51 | COMA | 70.59 | 61.52 | 68.61 | 0.45 | 0.39 | 0.19 | 70.59 | 61.52 | 68.61 | 0.29 | 0.17 | COMA | 51 | |
| 52 | Jones−UCL | 70.59 | 60.29 | 70.32 | 0.45 | 0.29 | 0.39 | 70.59 | 60.29 | 70.32 | 0.29 | 0.05 | 0.16 | Jones−UCL | 52 |
| 53 | nFOLD3 | 70.59 | 59.80 | 67.93 | 0.45 | 0.24 | 0.12 | 70.96 | 64.22 | 68.06 | 0.32 | 0.42 | nFOLD3 | 53 | |
| 54 | A−TASSER | 70.59 | 58.33 | 71.15 | 0.45 | 0.12 | 0.48 | 78.68 | 75.49 | 76.00 | 1.04 | 1.49 | 0.84 | A−TASSER | 54 |
| 55 | fleil | 70.22 | 63.85 | 68.85 | 0.42 | 0.59 | 0.22 | 72.06 | 64.46 | 71.52 | 0.43 | 0.45 | 0.31 | fleil | 55 |
| 56 | Chicken_George | 70.22 | 63.36 | 72.51 | 0.42 | 0.55 | 0.63 | 74.63 | 63.36 | 78.65 | 0.67 | 0.34 | 1.15 | Chicken_George | 56 |
| 57 | MUProt | 70.22 | 61.89 | 70.31 | 0.42 | 0.42 | 0.39 | 70.59 | 61.89 | 70.52 | 0.29 | 0.20 | 0.19 | MUProt | 57 |
| 58 | MULTICOM−CMFR | 70.22 | 61.64 | 70.13 | 0.42 | 0.40 | 0.37 | 72.43 | 65.56 | 71.51 | 0.46 | 0.55 | 0.30 | MULTICOM−CMFR | 58 |
| 59 | 3DShot2 | 70.22 | 61.40 | 68.45 | 0.42 | 0.38 | 0.18 | 70.22 | 61.40 | 68.45 | 0.25 | 0.16 | 3DShot2 | 59 | |
| 60 | HHpred4 | 70.22 | 61.15 | 70.24 | 0.42 | 0.36 | 0.38 | 70.22 | 61.15 | 70.24 | 0.25 | 0.13 | 0.15 | HHpred4 | 60 |
| 61 | HHpred5 | 70.22 | 61.15 | 70.24 | 0.42 | 0.36 | 0.38 | 70.22 | 61.15 | 70.24 | 0.25 | 0.13 | 0.15 | HHpred5 | 61 |
| 62 | MULTICOM−REFINE | 70.22 | 60.66 | 70.05 | 0.42 | 0.32 | 0.36 | 70.22 | 61.89 | 70.48 | 0.25 | 0.20 | 0.18 | MULTICOM−REFINE | 62 |
| 63 | MULTICOM−RANK | 70.22 | 60.42 | 70.17 | 0.42 | 0.30 | 0.37 | 72.06 | 63.97 | 70.66 | 0.43 | 0.40 | 0.20 | MULTICOM−RANK | 63 |
| 64 | 3DShot1 | 69.85 | 61.52 | 72.64 | 0.39 | 0.39 | 0.65 | 69.85 | 61.52 | 72.64 | 0.22 | 0.17 | 0.44 | 3DShot1 | 64 |
| 65 | MULTICOM−CLUSTER | 69.85 | 61.03 | 70.26 | 0.39 | 0.35 | 0.38 | 71.32 | 63.97 | 70.26 | 0.36 | 0.40 | 0.16 | MULTICOM−CLUSTER | 65 |
| 66 | PRI−Yang−KiharA | 69.85 | 58.58 | 70.15 | 0.39 | 0.14 | 0.37 | 69.85 | 58.58 | 70.15 | 0.22 | 0.14 | PRI−Yang−KiharA | 66 | |
| 67 | FAMSD | 69.85 | 54.90 | 69.30 | 0.39 | 0.27 | 69.85 | 62.38 | 69.30 | 0.22 | 0.25 | 0.04 | FAMSD | 67 | |
| 68 | SAM−T06−server | 69.48 | 61.89 | 72.26 | 0.36 | 0.42 | 0.61 | 69.48 | 62.74 | 72.26 | 0.19 | 0.28 | 0.39 | SAM−T06−server | 68 |
| 69 | pipe_int | 69.48 | 60.66 | 66.88 | 0.36 | 0.32 | 69.48 | 60.66 | 66.88 | 0.19 | 0.09 | pipe_int | 69 | ||
| 70 | Elofsson | 69.48 | 60.66 | 70.29 | 0.36 | 0.32 | 0.38 | 70.22 | 60.91 | 70.32 | 0.25 | 0.11 | 0.16 | Elofsson | 70 |
| 71 | HHpred2 | 69.48 | 60.66 | 69.66 | 0.36 | 0.32 | 0.31 | 69.48 | 60.66 | 69.66 | 0.19 | 0.09 | 0.08 | HHpred2 | 71 |
| 72 | SHORTLE | 69.48 | 54.53 | 75.43 | 0.36 | 0.96 | 69.48 | 54.53 | 75.43 | 0.19 | 0.77 | SHORTLE | 72 | ||
| 73 | FFASstandard | 69.12 | 62.50 | 67.72 | 0.33 | 0.47 | 0.09 | 69.12 | 63.97 | 67.72 | 0.15 | 0.40 | FFASstandard | 73 | |
| 74 | FUGUE_KM | 68.75 | 62.62 | 66.70 | 0.30 | 0.48 | 68.75 | 62.62 | 66.70 | 0.12 | 0.27 | FUGUE_KM | 74 | ||
| 75 | ABIpro | 68.75 | 60.17 | 67.42 | 0.30 | 0.28 | 0.06 | 76.47 | 71.08 | 77.58 | 0.84 | 1.07 | 1.03 | ABIpro | 75 |
| 76 | FFASsuboptimal | 68.38 | 54.90 | 67.66 | 0.27 | 0.09 | 71.69 | 62.62 | 70.62 | 0.39 | 0.27 | 0.20 | FFASsuboptimal | 76 | |
| 77 | TJ_Jiang | 68.02 | 59.44 | 67.37 | 0.24 | 0.21 | 0.05 | 69.12 | 60.17 | 70.52 | 0.15 | 0.04 | 0.19 | TJ_Jiang | 77 |
| 78 | FOLDpro | 68.02 | 59.19 | 68.23 | 0.24 | 0.19 | 0.15 | 68.02 | 59.19 | 68.23 | 0.05 | FOLDpro | 78 | ||
| 79 | mGenTHREADER | 68.02 | 56.01 | 66.22 | 0.24 | 68.02 | 56.01 | 66.22 | 0.05 | mGenTHREADER | 79 | ||||
| 80 | FFASflextemplate | 67.65 | 63.97 | 65.52 | 0.21 | 0.60 | 69.12 | 63.97 | 67.72 | 0.15 | 0.40 | FFASflextemplate | 80 | ||
| 81 | GS−MetaServer2 | 67.65 | 60.54 | 68.93 | 0.21 | 0.31 | 0.23 | 69.48 | 60.54 | 70.21 | 0.19 | 0.07 | 0.15 | GS−MetaServer2 | 81 |
| 82 | PSI | 67.28 | 62.87 | 64.88 | 0.18 | 0.50 | 67.28 | 62.87 | 64.88 | 0.30 | PSI | 82 | |||
| 83 | LOOPP_Server | 67.28 | 60.66 | 61.60 | 0.18 | 0.32 | 67.28 | 60.66 | 61.60 | 0.09 | LOOPP_Server | 83 | |||
| 84 | CpHModels | 66.91 | 62.99 | 64.96 | 0.15 | 0.51 | 66.91 | 62.99 | 64.96 | 0.31 | CpHModels | 84 | |||
| 85 | Pcons_local | 66.91 | 62.50 | 64.96 | 0.15 | 0.47 | 69.48 | 62.74 | 70.01 | 0.19 | 0.28 | 0.13 | Pcons_local | 85 | |
| 86 | Pcons_dot_net | 66.91 | 62.50 | 64.96 | 0.15 | 0.47 | 69.85 | 62.74 | 70.27 | 0.22 | 0.28 | 0.16 | Pcons_dot_net | 86 | |
| 87 | Pcons_multi | 66.91 | 62.50 | 64.96 | 0.15 | 0.47 | 69.85 | 62.99 | 68.76 | 0.22 | 0.31 | Pcons_multi | 87 | ||
| 88 | Softberry | 66.91 | 57.11 | 65.25 | 0.15 | 0.01 | 66.91 | 57.11 | 65.25 | Softberry | 88 | ||||
| 89 | GeneSilicoMetaServer | 66.54 | 59.68 | 66.71 | 0.12 | 0.23 | 69.48 | 60.54 | 70.21 | 0.19 | 0.07 | 0.15 | GeneSilicoMetaServer | 89 | |
| 90 | xianmingpan | 65.81 | 57.48 | 63.00 | 0.06 | 0.05 | 65.81 | 57.48 | 63.00 | xianmingpan | 90 | ||||
| 91 | SAM−T02−server | 65.81 | 57.23 | 63.94 | 0.06 | 0.03 | 65.81 | 57.23 | 63.94 | SAM−T02−server | 91 | ||||
| 92 | SMEG−CCP | 65.81 | 55.27 | 63.87 | 0.06 | 65.81 | 55.27 | 63.87 | SMEG−CCP | 92 | |||||
| 93 | JIVE08 | 65.07 | 53.31 | 63.52 | 0.00 | 65.07 | 55.15 | 63.52 | JIVE08 | 93 | |||||
| 94 | FLOUDAS | 57.72 | 43.51 | 54.58 | 57.72 | 43.51 | 54.58 | FLOUDAS | 94 | ||||||
| 95 | Handl−Lovell | 55.52 | 43.75 | 59.83 | 58.09 | 49.02 | 63.69 | Handl−Lovell | 95 | ||||||
| 96 | Sasaki−Cetin−Sasai | 54.04 | 49.39 | 60.14 | 54.04 | 49.39 | 60.14 | Sasaki−Cetin−Sasai | 96 | ||||||
| 97 | SAINT1 | 51.84 | 41.79 | 59.92 | 51.84 | 41.79 | 59.92 | SAINT1 | 97 | ||||||
| 98 | FALCON_CONSENSUS | 49.27 | 40.20 | 49.27 | 50.73 | 40.20 | 49.27 | FALCON_CONSENSUS | 98 | ||||||
| 99 | FALCON | 48.90 | 43.75 | 58.60 | 49.27 | 43.75 | 58.60 | FALCON | 99 | ||||||
| 100 | keasar | 44.12 | 42.28 | 56.43 | 73.90 | 63.85 | 77.31 | 0.60 | 0.39 | 0.99 | keasar | 100 | |||
| 101 | fais@hgc | 43.75 | 37.87 | 58.01 | 54.04 | 44.12 | 60.88 | fais@hgc | 101 | ||||||
| 102 | RBO−Proteus | 43.38 | 41.79 | 55.89 | 43.38 | 41.79 | 58.06 | RBO−Proteus | 102 | ||||||
| 103 | MUFOLD−MD | 40.44 | 34.31 | 50.33 | 50.00 | 40.32 | 55.44 | MUFOLD−MD | 103 | ||||||
| 104 | POEMQA | 40.44 | 33.09 | 57.14 | 75.73 | 66.05 | 80.58 | 0.77 | 0.60 | 1.38 | POEMQA | 104 | |||
| 105 | forecast | 40.07 | 29.53 | 52.46 | 40.81 | 33.46 | 52.46 | forecast | 105 | ||||||
| 106 | PHAISTOS | 39.71 | 36.77 | 51.55 | 58.09 | 48.77 | 56.23 | PHAISTOS | 106 | ||||||
| 107 | Jiang_Zhu | 39.34 | 34.93 | 49.85 | 39.34 | 36.64 | 49.85 | Jiang_Zhu | 107 | ||||||
| 108 | fais−server | 39.34 | 33.21 | 56.36 | 49.27 | 46.57 | 57.24 | fais−server | 108 | ||||||
| 109 | circle | 38.60 | 34.44 | 45.35 | 71.69 | 63.85 | 68.95 | 0.39 | 0.39 | 0.00 | circle | 109 | |||
| 110 | dill_ucsf | 38.60 | 32.97 | 43.69 | 38.60 | 32.97 | 43.69 | dill_ucsf | 110 | ||||||
| 111 | Frankenstein | 38.60 | 31.00 | 48.10 | 45.59 | 36.89 | 48.10 | Frankenstein | 111 | ||||||
| 112 | FrankensteinLong | 38.60 | 31.00 | 48.10 | 45.59 | 36.89 | 48.10 | FrankensteinLong | 112 | ||||||
| 113 | RPFM | 38.60 | 28.43 | 42.15 | 38.60 | 33.46 | 48.62 | RPFM | 113 | ||||||
| 114 | Wolynes | 38.23 | 30.15 | 46.72 | 42.65 | 37.87 | 46.90 | Wolynes | 114 | ||||||
| 115 | DistillSN | 37.87 | 26.59 | 33.40 | 41.91 | 36.52 | 38.31 | DistillSN | 115 | ||||||
| 116 | MeilerLabRene | 37.50 | 37.01 | 54.90 | 40.44 | 37.26 | 57.75 | MeilerLabRene | 116 | ||||||
| 117 | Distill | 36.40 | 30.02 | 49.70 | 40.44 | 32.11 | 51.08 | Distill | 117 | ||||||
| 118 | Scheraga | 35.66 | 26.59 | 29.87 | 36.40 | 31.98 | 37.28 | Scheraga | 118 | ||||||
| 119 | igor | 34.93 | 31.00 | 37.45 | 34.93 | 31.00 | 37.45 | igor | 119 | ||||||
| 120 | ShakAbInitio | 34.19 | 26.59 | 44.72 | 38.23 | 29.17 | 44.72 | ShakAbInitio | 120 | ||||||
| 121 | mariner1 | 34.19 | 24.63 | 27.53 | 39.34 | 30.39 | 48.59 | mariner1 | 121 | ||||||
| 122 | POEM | 33.82 | 25.49 | 50.69 | 43.75 | 40.44 | 80.58 | 1.38 | POEM | 122 | |||||
| 123 | MUFOLD−Server | 33.09 | 26.72 | 43.02 | 39.34 | 33.21 | 48.72 | MUFOLD−Server | 123 | ||||||
| 124 | rehtnap | 31.98 | 29.78 | 32.60 | 31.98 | 29.78 | 32.60 | rehtnap | 124 | ||||||
| 125 | schenk−torda−server | 31.62 | 25.25 | 31.01 | 33.82 | 27.45 | 32.21 | schenk−torda−server | 125 | ||||||
| 126 | RANDOM | 29.00 | 23.84 | 29.00 | 29.00 | 23.84 | 29.00 | RANDOM | 126 | ||||||
| 127 | Linnolt−UH−CMB | 26.47 | 22.30 | 33.97 | 26.47 | 22.30 | 33.97 | Linnolt−UH−CMB | 127 | ||||||
| 128 | panther_server | 26.10 | 24.39 | 13.56 | 26.10 | 24.39 | 13.56 | panther_server | 128 | ||||||
| 129 | mahmood−torda−server | 25.00 | 21.20 | 38.90 | 29.04 | 26.23 | 38.90 | mahmood−torda−server | 129 | ||||||
| 130 | Pushchino | 23.90 | 21.69 | 7.77 | 23.90 | 21.69 | 7.77 | Pushchino | 130 | ||||||
| 131 | TWPPLAB | 22.79 | 19.61 | 8.14 | 22.79 | 19.61 | 8.14 | TWPPLAB | 131 | ||||||
| 132 | DelCLab | 22.43 | 19.73 | 8.17 | 23.90 | 20.59 | 15.25 | DelCLab | 132 | ||||||
| 133 | 3Dpro | 3Dpro | 133 | ||||||||||||
| 134 | Abagyan | Abagyan | 134 | ||||||||||||
| 135 | BHAGEERATH | BHAGEERATH | 135 | ||||||||||||
| 136 | CBSU | CBSU | 136 | ||||||||||||
| 137 | FEIG_REFINE | FEIG_REFINE | 137 | ||||||||||||
| 138 | Fiser−M4T | Fiser−M4T | 138 | ||||||||||||
| 139 | HCA | HCA | 139 | ||||||||||||
| 140 | LEE−SERVER | LEE−SERVER | 140 | ||||||||||||
| 141 | NIM2 | NIM2 | 141 | ||||||||||||
| 142 | Nano_team | Nano_team | 142 | ||||||||||||
| 143 | OLGAFS | OLGAFS | 143 | ||||||||||||
| 144 | Ozkan−Shell | Ozkan−Shell | 144 | ||||||||||||
| 145 | POISE | POISE | 145 | ||||||||||||
| 146 | PZ−UAM | PZ−UAM | 146 | ||||||||||||
| 147 | ProtAnG | ProtAnG | 147 | ||||||||||||
| 148 | ProteinShop | ProteinShop | 148 | ||||||||||||
| 149 | SAMUDRALA | SAMUDRALA | 149 | ||||||||||||
| 150 | TsaiLab | TsaiLab | 150 | ||||||||||||
| 151 | UCDavisGenome | UCDavisGenome | 151 | ||||||||||||
| 152 | Wolfson−FOBIA | Wolfson−FOBIA | 152 | ||||||||||||
| 153 | YASARA | YASARA | 153 | ||||||||||||
| 154 | YASARARefine | YASARARefine | 154 | ||||||||||||
| 155 | Zhou−SPARKS | Zhou−SPARKS | 155 | ||||||||||||
| 156 | dill_ucsf_extended | dill_ucsf_extended | 156 | ||||||||||||
| 157 | huber−torda−server | huber−torda−server | 157 | ||||||||||||
| 158 | jacobson | jacobson | 158 | ||||||||||||
| 159 | mti | mti | 159 | ||||||||||||
| 160 | mumssp | mumssp | 160 | ||||||||||||
| 161 | psiphifoldings | psiphifoldings | 161 | ||||||||||||
| 162 | ricardo | ricardo | 162 | ||||||||||||
| 163 | rivilo | rivilo | 163 | ||||||||||||
| 164 | sessions | sessions | 164 | ||||||||||||
| 165 | taylor | taylor | 165 | ||||||||||||
| 166 | test_http_server_01 | test_http_server_01 | 166 | ||||||||||||