T0493
C-terminal domain of DNA helicase APC89291.2 from Lactobacillus plantarun
Target sequence:
>T0493 DNA helicase APC89291.2, Lactobacillus plantarun, 174 residues
SNASYRSTQQITDFTKEILVNGEAVTAFDRQGDLPNVVVTPNFEAGVDQVVDQLAMNDSERDTTAIIGKSLAECEALTKALKARGEQVTLIQTENQRLAPGVIVVPSFLAKGLEFDAVIVWNANQENYQREDERQLLYTICSRAMHELTLVAVGSLSPLLARVNHALYTLNEAK
Structure:
Determined by:
MCSG
PDB ID: 3dmn
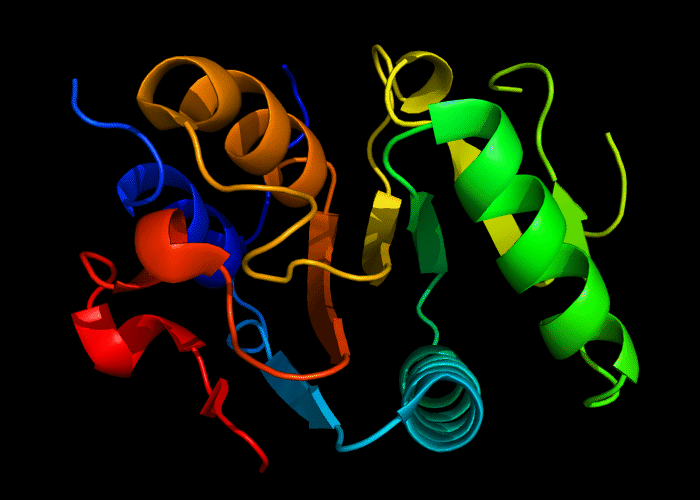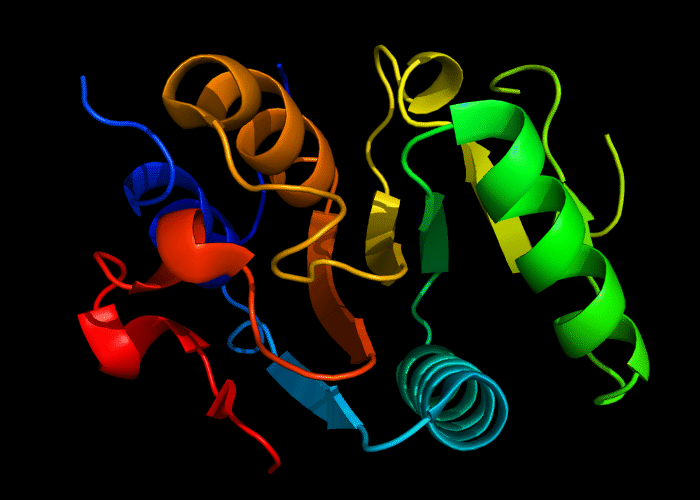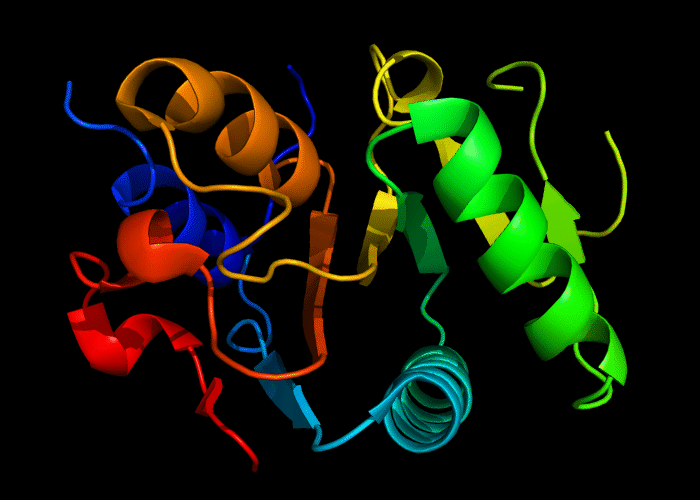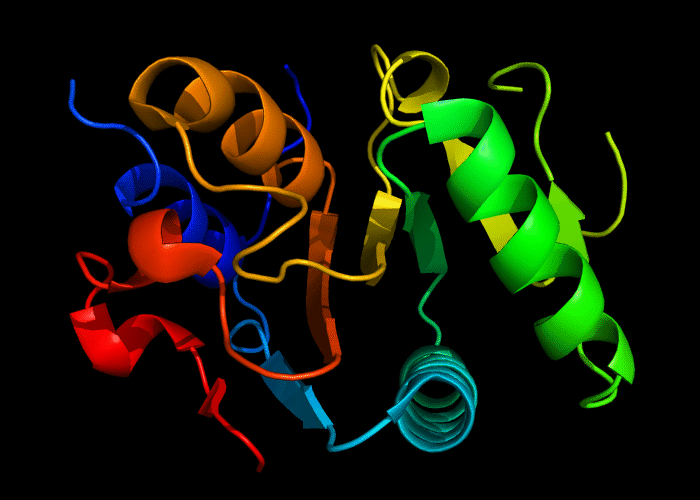
Cartoon diagram of 493: 3dmn
Domains: PyMOL of domains
Single domain. A piece of a larger protein, which is nevetheless stable and structured, except two loops.
Structure classification:
P-loop-containing nucleoside triphosphate hydrolase fold, P-loop motif deteriorated.
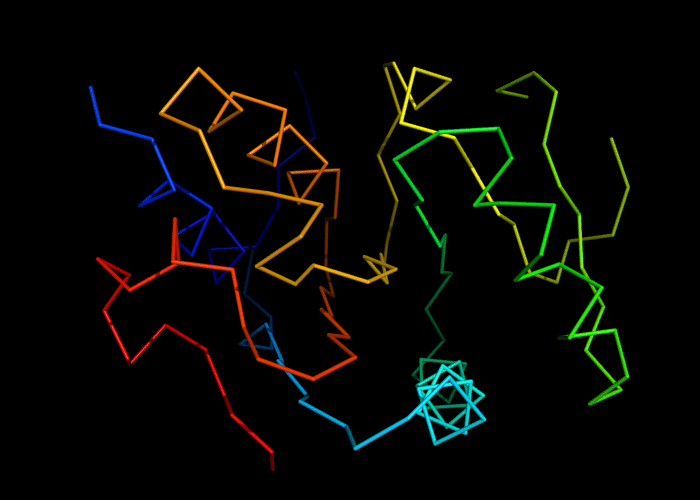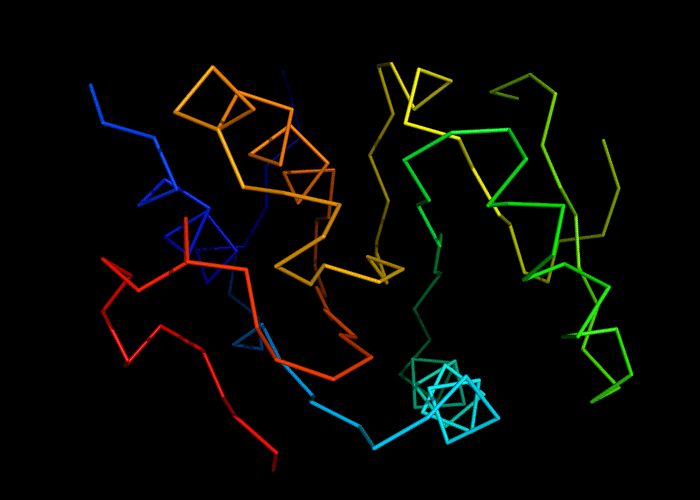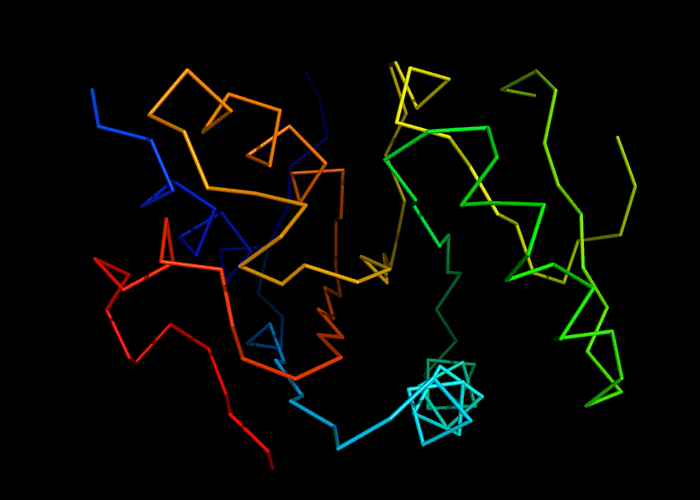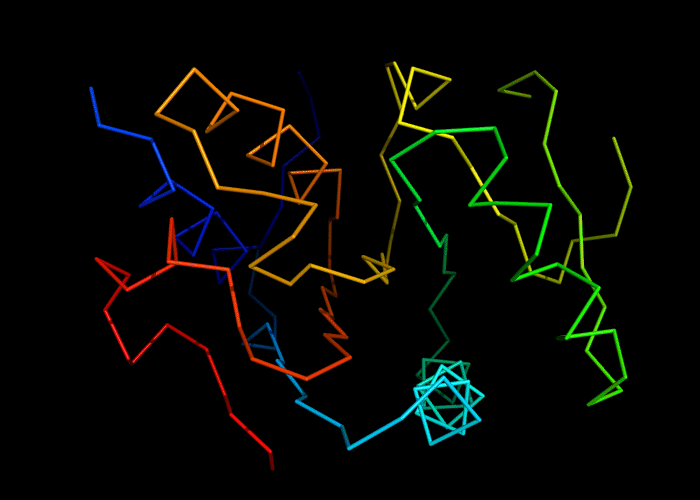
Ribbon diagram of 493: 3dmn
CASP category:
Comparative modeling:medium.
Closest templates:
Conservation: 8 69978 58 5 5 9 6 5 665 7 5 5 6 6 5 5997997 7 5 85 7976 7 7 5 2IS1_A_ -IRLEQNYRSTSNILSAANALIENNNGR------LGKKLWTDGADGEPISLYCAFNELDEARFVVNRIKTWQDNGGALAECAILYRSNAQSRVLEEALLQASMPYRIYGGMRFFERQEIEGQADT-WQDAVQLMTLHSAKGLEFPQVFIVGMEEGMFPSQMSL--------DEGGRLEEERRLAYVGVTRAMQKLTLTYAETRRLYGKEVYHRPSRFIGELPEECVEEVRLRATVSRPVSHQRMGTPMVENDSGYKLGQRVRHAK 174
gi_108797788_ref_YP_63798 SSRLNINYRTTAEILGWSLGLMRGEPIDDMEGGLDSIAGCKSYVHGSPPMLSGSGTADAEAEHIAKVVNAWIDRGVAPSEIGIAVRAKWFASKIEHALKAAGIAT------------VDLARASE-DDEAVHVGTMHRMKGLEFRCMCVAGVTAKWVPALNAVTPIDDDKQTHQRDLERERCLLFVACTRAREELLVTWHGE-----------PSPFIADLAHGKEENQ------------------------------------ 675
gi_168214788_ref_ZP_02640 IYNLNKSYRSTYEIMEYANKYLDED------------RIIPIVRHGKPVEEIEFHNDEELSESIIESLKEFSNEG--LESIAIITRDKEELEKVYNLISNKVHL------------VKFDNEDVL-YKGGNVIIPSYFAKGLEFDGVIIVDNGSLKD--------------------ENEDLIKYIMSTRALHRLKEIQFK---------------------------------------------------------------- 695
gi_169191099_ref_ZP_02850 FFELNRSYRSTMEIIEFANRILGAMGGG-------VKPAVPVFRSGDPVEVVQVDRAAWQKQIAQSIREWQADGQ--FQTISVIGRTAEQCAELHDYLVREGLEA-----------SLVQSKQPE-YGGGLSVVPVYLSKGLEFDAVLIADAGADMYT-------------------QDDAKLLYVGCTRALHKLKLLHRES-----------LTTLINEV-------------------------------------------- 544
gi_168158683_ref_ZP_02593 YYELTRSYRSTKEIIEFANEIIKNAEIP-------VGLATPVFRSGEEVKVIQAEDQFNEIMKTLKHLQNDD-----VKTIAVIGRTDDECRDIYEKLTKAGVAV-----------NVIEADQSK-YEGGISVVPVYLAKGLEFDAVLLIDVDEEHYKN-----------------TKHDAKLLYVGCTRSLHDLWIFHSGE-----------ASPLIKK--------------------------------------------- 670
gi_168165560_ref_ZP_02600 YYELTRSYRSTKEIIEFANEIIKNAEIP-------VGLAMPVFRSGEEVKVIHTEDQFTEILKTLKQLQNAD-----VKTIAVIGRTDDECRDMYEKLTNAGLTV-----------NVIEANQSK-YEGGISVVPVYLAKGLEFDAVLLIDVDEEHYKN-----------------TKHDAKLLYVGCTRSLHDLWIFHSGE-----------ASPLIKK--------------------------------------------- 670gi_169826457_ref_YP_00169 FRTLQKSYRTTIEIMEVANEILAQMDEQ-------LPLVEPVVRHGNVPSFIQAEQFDAL--KIKEIFDTIRQHG--HRSIALICKTTVEAIMMQNDLTAHQLPA-----------QLLTENDSI-NQEMLLVVPSHLAKGLEFDAVIVAAFHTPFYD------------------TPIDRKLLYVALTRAMHELYLIGPSK-----------NTFLLEN--------------------------------------------- 723
gi_110799377_ref_YP_69630 YKHLSQSYRSTVEIINYANKVLVKQNTY-------EEPAKPVLRHGMVPEEIEFNNAKEFCERLDNIVKKVKDEG--KTSIAIVGKTLDECKKIRTLVKKYSNYD----------FELIKDDQKE-FDLNLIIIPSYLTKGLEFDCTIVYNLNKENYND-----------------NEIDKKLLYVVLTRALHYEYIFYKEE-----------KSPLIG---------------------------------------------- 745
gi_29375517_ref_NP_814671 TYQLLNSYRSSKEITRLFQTLGTQPEQ---------LTIVPIRHDGEKPTFIPCDSASDYLKAIVTSITTFDSK----ESTAIITKTFAEKEQLTKELCA--------------------SLSLE-EHPSLQVLSIDMAKGLEFDNVILHNPNVNRYSD-----------------NKRDKKILYTAISRGMKRVVLPYTGT-----------LSEWFTPYLAAQ---------------------------------------- 708
gi_28377335_ref_NP_784227 LITLNKSYRSTEQITNFAKALLPDGD-----------QIQAFTRKGDLPKVMIRYGEDAALSSLRSEVEHQLKA---TNTVAVLTKDLAESKQVYQYLKRY-TVT-----------TLMADSDRT-MPQGVIVMPIYLAKGLEFDAVVAYDVSATNYP------------------DADSVGILYTIASRAMHHLTLLSIGD-----------VSPLIAQLDPTLFTIEHQVRV------------------------------- 736
gi_148543438_ref_YP_00127 LIALRRSYRSTTEITNFAKALLPDGD-----------KIISFTRHGKKPRLLVRYSDKESQQSLLDETLKLADE---HETVAILTKNQEQAAGIYQLLHRQKVEN-----------IHLLDKDASELPKGVLILPIYLAKGLEFDAVLAADVSAKNLT------------------NTDEVGMIYTMASRAMHELVLLSNGS-----------VSDAINEKAGRLLTIEYQLPNKN----------------------------- 737gi_116492059_ref_YP_80379 LITLNRSYRSTAQITNFAAALLPDGH-----------KIQAFSREGDLPKVIIADNEQQAVVDMTNLCRDELQQ---YGTIAILTKNMAESEHVATLLHELKP-------------SLLNDGDRS-LPKGIVVLPIYLAKGLEFDSVIAYNVSEANFP------------------GRLWRGTLYTIASRAMHQLHLISIGK-----------VSNLITQIKPDQFSFQKSFNITPHKSQKLDE--------------------- 724gi_42519709_ref_NP_965639 VVQLTKSYRSTKEITNFTKQILRQGE-----------KIEAFDRRGPKPAFYKRDSIEKEYSALEDILAENDEQ---KLTTAIITKTLAEAQEVAKVLKERKIKA-----------TLIGSANQR-LVPGTLVMPSYLAKGLEFDAVVAWGVSKENYH------------------QLDETQLLYTIASRAMYKLDLIYTGE-----------KSPLFDNLDEKTYVNK------------------------------------ 736
gi_116495451_ref_YP_80718 VVQLTQSYRSTKQVTDFTKAILTSGQ-----------KIVAFNRQGPLPKLVVRPDEAGLMAALQDQLKINNEA---KQTTAVIAKTLEAAEAIYEQMKAAGIKV-----------TLIKSENQR-LAAGVIVVPSFLAKGLEFDAVVLWQINAANYH------------------EEDERELLYTVASRAMHRLTMLASPD-----------VTPLLDKVPADLYERG------------------------------------ 735T0493_DNA_helicase_APC892 ---SNASYRSTQQITDFTKEILVNGE-----------AVTAFDRQGDLPNVVVTPNFEAGVDQVVDQLAMNDSE---RDTTAIIGKSLAECEALTKALKARGEQV-----------TLIQTENQR-LAPGVIVVPSFLAKGLEFDAVIVWNANQENYQ------------------REDERQLLYTICSRAMHELTLVAVGS-----------LSPLLARVNHALYTLNEAK--------------------------------- 143
gi_168135096_ref_ZP_02578 GINLTRSYRSTKPIIEFTRALVPEGK-----------NIHAFERDGEKPTVTKVSNESELHEHITAKVAELQKEQ--HNTIAIICKSAAESAAAYEALNHI-ENI-----------KLVKSNSAE-YEQGIVVIPAYLAKGIEFDAVIIYDASKDVYS------------------DESVRRLFYTACTRAMHELHLYSVGE-----------VSPFVLGADSESFELITTP--------------------------------- 746
gi_49480066_ref_YP_035300 GINLTRSYRSTKPIIEFTRALVPEGK-----------NIHAFERDGEKPTVTKVSNHNELHEHITSKIAELQKQN--HNTIAIICKSAAESAAAYEALSHI-ENI-----------KLVKSNSAE-YEQGIVVIPAYLAKGIEFDAVIIYDASKDVYN------------------DESVRRLFYTACTRAMHELQLYSVGE-----------VSPFIVEADSESFELIT-P--------------------------------- 746gi_167635442_ref_ZP_02393 GINLTRSYRSTKPIIEFTRALVPEGK-----------NIHAFERDGEKPTVTKVANESELHERITAKVAELQKQN--HNTIAIICKSAAESAAAYEALSPI-ENI-----------KLVKSNSAE-YEQGIVVIPAYLAKGIEFDAVIIYDASKAVYN------------------DESVRRLFYTACTRAMHELQLYSVGE-----------VSPFIVEADSESFELIT-P--------------------------------- 746gi_16080398_ref_NP_391225 YVRLKRTYRSTRQIVEFTKAMLQDGA-----------DIEPFNRSGEMPLVVKTEGHESLCQKLAQEIGRLKKKG--HETIAVICKTAHQCIQAHAHMSEY-TDV-----------RLIHKENQP-FQKGVCVIPVYLAKGIEFDAVLVYDASEEHYH------------------TEHDRRLLYTACTRAMHMLAVFYTGE-----------ASPFVTAVPPHLYQIAE----------------------------------- 745
gi_89895484_ref_YP_518971 GMELCRSYRSTIEISGFTQKILENK------------KLIPIERHGDTPTITVCRTPKEQLAEIQHLIEEFRQSE--YATLGVICKSQQQARLLFAQLHGVYDTM-----------VLLDFGSSE-FQEGIVVTSVHMAKGLEFDQVIVPEVSDDLFK------------------TQLDRSLLYIACTRAMHKLDLTCCGE-----------KTKFLHSC-------------------------------------------- 274
Consensus_aa: ..pLppoYRST.pIhphhp.ll.p..............h.sh.+pGp.sphh...s...bh..l.p.h....pp.....ohAlls+s..pt..h.p.lp.....h............hh.s.s.p.b..tl.lhs.@hAKGLEFDsVllhshs...h....................p.-..lhYhhhoRAhccL.lh...p............o.hl.p.............................................Consensus_ss: eeee hhhhhhhhhhhhh eeeeee hhhhhhhhhhhhhhhhh eeeee hhhhhhhhhhhhh hhh h eeee hhh eeeee hhhhhhhhhhhhhh eeeeee hhhhh hhh Target sequence - PDB file inconsistencies:
T0493 3dmn.pdb T0493.pdb PyMOL PyMOL of domains
T0493 1 SNASYRSTQQITDFTKEILVNGEAVTAFDRQGDLPNVVVTPNFEAGVDQVVDQLAMNDSERDTTAIIGKSLAECEALTKALKARGEQVTLIQTENQRLAPGVIVVPSFLAKGLEFDAVIVWNANQENYQREDERQLLYTICSRAMHELTLVAVGSLSPLLARVNHALYTLNEAK 174 ~~|||||||||||||||||||~~~~~~~~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~ 3dmnA 596 --ASYRSTQQITDFTKEILVN--------RQGDLPNVVVTPNFEAGVDQVVDQLAMNDSERDTTAIIGKSLAECEALTKALKARGEQVTLIQTEN-RLAPGVIVVPSFLAKGLEFDAVIVWNANQENYQREDERQLLYTICSRAMHELTLVAVGSLSPLLARVNHALYTLNE-- 765
Residue change log: change 649, 738, MSE to MET;
Single domain protein: target 3-21, 30-95, 97-172 ; pdb 596-614, 623-688, 690-765
Sequence classification:
A domain from COG3973 in CDD.
Server predictions:
T0493:pdb 596-765:seq 3-172:CM_medium; alignment
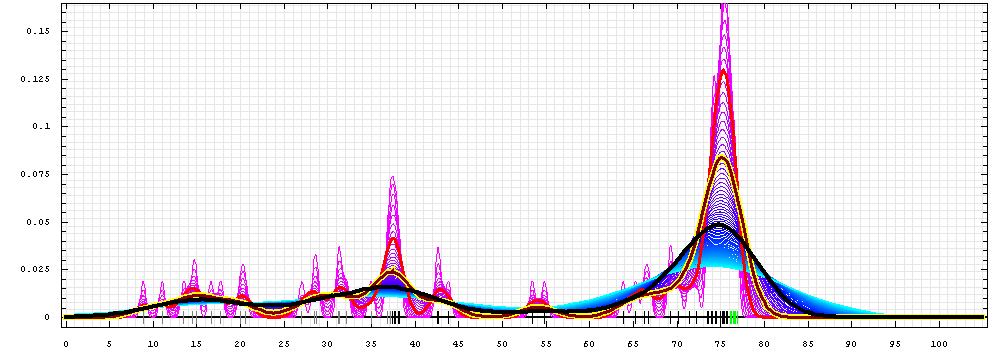
First models for T0493:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0493 | T0493 | T0493 | T0493 | ||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||
| 1 | EB_AMU_Physics | 77.64 | 72.88 | 78.10 | 0.80 | 0.86 | 0.74 | 77.64 | 72.88 | 78.75 | 0.74 | 0.77 | 0.70 | EB_AMU_Physics | 1 |
| 2 | IBT_LT | 77.17 | 72.62 | 77.93 | 0.77 | 0.85 | 0.74 | 77.17 | 72.62 | 77.93 | 0.71 | 0.76 | 0.65 | IBT_LT | 2 |
| 3 | LEE | 77.17 | 71.07 | 81.06 | 0.77 | 0.76 | 0.91 | 77.33 | 72.46 | 81.06 | 0.72 | 0.75 | 0.84 | LEE | 3 |
| 4 | MULTICOM−RANK | 76.86 | 71.79 | 77.77 | 0.75 | 0.80 | 0.73 | 76.86 | 71.79 | 78.19 | 0.69 | 0.70 | 0.67 | MULTICOM−RANK | 4 |
| 5 | KudlatyPredHuman | 76.86 | 71.58 | 78.14 | 0.75 | 0.79 | 0.75 | 76.86 | 71.58 | 78.84 | 0.69 | 0.69 | 0.71 | KudlatyPredHuman | 5 |
| 6 | MULTICOM | 76.86 | 70.76 | 78.54 | 0.75 | 0.74 | 0.77 | 77.02 | 72.41 | 78.54 | 0.70 | 0.74 | 0.69 | MULTICOM | 6 |
| 7 | 3DShotMQ | 76.71 | 71.64 | 78.13 | 0.75 | 0.79 | 0.75 | 76.71 | 71.64 | 78.13 | 0.68 | 0.69 | 0.66 | 3DShotMQ | 7 |
| 8 | POEMQA | 76.71 | 71.53 | 77.85 | 0.75 | 0.79 | 0.73 | 76.71 | 71.53 | 78.45 | 0.68 | 0.69 | 0.68 | POEMQA | 8 |
| 9 | MUProt | 76.71 | 71.53 | 78.60 | 0.75 | 0.79 | 0.77 | 77.02 | 71.58 | 78.60 | 0.70 | 0.69 | 0.69 | MUProt | 9 |
| 10 | Elofsson | 76.71 | 71.43 | 78.13 | 0.75 | 0.78 | 0.75 | 77.02 | 71.64 | 78.68 | 0.70 | 0.69 | 0.70 | Elofsson | 10 |
| 11 | Frankenstein | 76.55 | 72.41 | 77.23 | 0.74 | 0.84 | 0.70 | 76.55 | 72.41 | 77.23 | 0.67 | 0.74 | 0.61 | Frankenstein | 11 |
| 12 | Zico | 76.55 | 71.58 | 77.86 | 0.74 | 0.79 | 0.73 | 76.55 | 71.95 | 79.51 | 0.67 | 0.71 | 0.75 | Zico | 12 |
| 13 | ZicoFullSTP | 76.55 | 71.58 | 77.86 | 0.74 | 0.79 | 0.73 | 76.55 | 71.95 | 79.51 | 0.67 | 0.71 | 0.75 | ZicoFullSTP | 13 |
| 14 | MULTICOM−REFINE | 76.55 | 70.34 | 78.21 | 0.74 | 0.72 | 0.75 | 77.17 | 71.89 | 78.64 | 0.71 | 0.71 | 0.69 | MULTICOM−REFINE | 14 |
| 15 | GeneSilico | 76.55 | 70.34 | 78.21 | 0.74 | 0.72 | 0.75 | 76.71 | 72.77 | 79.28 | 0.68 | 0.77 | 0.73 | GeneSilico | 15 |
| 16 | McGuffin | 76.55 | 70.34 | 78.21 | 0.74 | 0.72 | 0.75 | 76.71 | 72.77 | 78.64 | 0.68 | 0.77 | 0.69 | McGuffin | 16 |
| 17 | GS−KudlatyPred | 76.40 | 72.77 | 76.97 | 0.73 | 0.86 | 0.68 | 76.40 | 72.77 | 77.03 | 0.66 | 0.77 | 0.60 | GS−KudlatyPred | 17 |
| 18 | MULTICOM−CMFR | 76.24 | 72.00 | 77.02 | 0.72 | 0.81 | 0.69 | 76.40 | 72.00 | 78.42 | 0.66 | 0.72 | 0.68 | MULTICOM−CMFR | 18 |
| 19 | Bates_BMM | 76.24 | 71.48 | 78.43 | 0.72 | 0.78 | 0.76 | 76.24 | 71.48 | 78.43 | 0.65 | 0.68 | 0.68 | Bates_BMM | 19 |
| 20 | mufold | 76.09 | 71.74 | 78.56 | 0.71 | 0.80 | 0.77 | 76.09 | 71.74 | 79.11 | 0.64 | 0.70 | 0.72 | mufold | 20 |
| 21 | MULTICOM−CLUSTER | 76.09 | 70.91 | 78.22 | 0.71 | 0.75 | 0.75 | 76.09 | 70.91 | 79.08 | 0.64 | 0.65 | 0.72 | MULTICOM−CLUSTER | 21 |
| 22 | Chicken_George | 75.93 | 69.72 | 76.08 | 0.70 | 0.68 | 0.63 | 76.71 | 72.77 | 78.81 | 0.68 | 0.77 | 0.70 | Chicken_George | 22 |
| 23 | TASSER | 75.93 | 67.03 | 78.93 | 0.70 | 0.53 | 0.79 | 75.93 | 71.22 | 78.93 | 0.63 | 0.67 | 0.71 | TASSER | 23 |
| 24 | 3Dpro | 75.78 | 71.22 | 75.55 | 0.69 | 0.77 | 0.60 | 77.33 | 73.71 | 76.87 | 0.72 | 0.83 | 0.59 | 3Dpro | 24 |
| 25 | FFASstandard | 75.78 | 70.60 | 76.16 | 0.69 | 0.73 | 0.64 | 75.78 | 70.60 | 76.41 | 0.62 | 0.63 | 0.56 | FFASstandard | 25 |
| 26 | FFASflextemplate | 75.78 | 70.60 | 76.16 | 0.69 | 0.73 | 0.64 | 75.78 | 70.60 | 76.16 | 0.62 | 0.63 | 0.54 | FFASflextemplate | 26 |
| 27 | MUSTER | 75.78 | 69.15 | 77.88 | 0.69 | 0.65 | 0.73 | 75.78 | 69.15 | 77.88 | 0.62 | 0.53 | 0.65 | MUSTER | 27 |
| 28 | HHpred5 | 75.62 | 71.07 | 77.94 | 0.68 | 0.76 | 0.74 | 75.62 | 71.07 | 77.94 | 0.61 | 0.66 | 0.65 | HHpred5 | 28 |
| 29 | FrankensteinLong | 75.62 | 70.34 | 76.06 | 0.68 | 0.72 | 0.63 | 75.62 | 70.34 | 76.15 | 0.61 | 0.61 | 0.54 | FrankensteinLong | 29 |
| 30 | Zhang−Server | 75.62 | 69.20 | 79.28 | 0.68 | 0.65 | 0.81 | 75.93 | 69.67 | 79.84 | 0.63 | 0.57 | 0.77 | Zhang−Server | 30 |
| 31 | Sternberg | 75.62 | 69.20 | 79.28 | 0.68 | 0.65 | 0.81 | 75.62 | 69.20 | 79.28 | 0.61 | 0.54 | 0.73 | Sternberg | 31 |
| 32 | Zhang | 75.62 | 68.48 | 80.50 | 0.68 | 0.61 | 0.88 | 75.78 | 70.76 | 80.59 | 0.62 | 0.64 | 0.81 | Zhang | 32 |
| 33 | pro−sp3−TASSER | 75.47 | 71.22 | 78.33 | 0.68 | 0.77 | 0.76 | 75.47 | 71.22 | 78.94 | 0.60 | 0.67 | 0.71 | pro−sp3−TASSER | 33 |
| 34 | fams−ace2 | 75.47 | 70.39 | 77.45 | 0.68 | 0.72 | 0.71 | 75.62 | 71.17 | 77.72 | 0.61 | 0.66 | 0.64 | fams−ace2 | 34 |
| 35 | METATASSER | 75.31 | 70.55 | 78.62 | 0.67 | 0.73 | 0.77 | 75.31 | 70.55 | 78.62 | 0.59 | 0.62 | 0.69 | METATASSER | 35 |
| 36 | COMA−M | 75.31 | 70.44 | 77.99 | 0.67 | 0.72 | 0.74 | 75.31 | 70.44 | 78.17 | 0.59 | 0.62 | 0.66 | COMA−M | 36 |
| 37 | COMA | 75.31 | 70.44 | 77.99 | 0.67 | 0.72 | 0.74 | 75.31 | 70.44 | 78.17 | 0.59 | 0.62 | 0.66 | COMA | 37 |
| 38 | forecast | 75.31 | 69.31 | 77.86 | 0.67 | 0.66 | 0.73 | 75.47 | 70.70 | 78.46 | 0.60 | 0.63 | 0.68 | forecast | 38 |
| 39 | POEM | 75.31 | 69.10 | 75.34 | 0.67 | 0.65 | 0.59 | 75.93 | 70.08 | 78.45 | 0.63 | 0.59 | 0.68 | POEM | 39 |
| 40 | 3DShot1 | 75.31 | 69.00 | 77.89 | 0.67 | 0.64 | 0.73 | 75.31 | 69.00 | 77.89 | 0.59 | 0.52 | 0.65 | 3DShot1 | 40 |
| 41 | ZicoFullSTPFullData | 75.31 | 68.38 | 79.13 | 0.67 | 0.61 | 0.80 | 76.55 | 71.58 | 79.51 | 0.67 | 0.69 | 0.75 | ZicoFullSTPFullData | 41 |
| 42 | HHpred4 | 75.16 | 69.56 | 77.58 | 0.66 | 0.67 | 0.72 | 75.16 | 69.56 | 77.58 | 0.58 | 0.56 | 0.63 | HHpred4 | 42 |
| 43 | ABIpro | 75.16 | 69.56 | 75.11 | 0.66 | 0.67 | 0.58 | 75.16 | 69.56 | 75.11 | 0.58 | 0.56 | 0.48 | ABIpro | 43 |
| 44 | Pcons_local | 75.00 | 70.65 | 75.88 | 0.65 | 0.74 | 0.62 | 75.00 | 70.65 | 75.88 | 0.57 | 0.63 | 0.53 | Pcons_local | 44 |
| 45 | Pcons_dot_net | 75.00 | 70.65 | 75.88 | 0.65 | 0.74 | 0.62 | 75.00 | 70.65 | 77.43 | 0.57 | 0.63 | 0.62 | Pcons_dot_net | 45 |
| 46 | Pcons_multi | 75.00 | 70.65 | 75.88 | 0.65 | 0.74 | 0.62 | 75.31 | 72.31 | 77.88 | 0.59 | 0.74 | 0.65 | Pcons_multi | 46 |
| 47 | MidwayFolding | 74.84 | 68.94 | 75.46 | 0.64 | 0.64 | 0.60 | 75.16 | 70.91 | 76.02 | 0.58 | 0.65 | 0.53 | MidwayFolding | 47 |
| 48 | FAMS−multi | 74.53 | 70.29 | 75.32 | 0.62 | 0.72 | 0.59 | 74.53 | 70.29 | 75.58 | 0.54 | 0.61 | 0.51 | FAMS−multi | 48 |
| 49 | Bilab−UT | 74.53 | 69.77 | 77.95 | 0.62 | 0.69 | 0.74 | 75.16 | 69.77 | 77.95 | 0.58 | 0.57 | 0.65 | Bilab−UT | 49 |
| 50 | RAPTOR | 74.53 | 69.15 | 76.12 | 0.62 | 0.65 | 0.64 | 74.53 | 69.20 | 76.12 | 0.54 | 0.54 | 0.54 | RAPTOR | 50 |
| 51 | keasar−server | 74.38 | 70.03 | 76.14 | 0.62 | 0.70 | 0.64 | 74.38 | 70.03 | 77.56 | 0.53 | 0.59 | 0.63 | keasar−server | 51 |
| 52 | FFASsuboptimal | 74.38 | 69.10 | 75.39 | 0.62 | 0.65 | 0.60 | 74.38 | 69.10 | 75.39 | 0.53 | 0.53 | 0.50 | FFASsuboptimal | 52 |
| 53 | circle | 74.38 | 67.86 | 77.35 | 0.62 | 0.58 | 0.70 | 76.24 | 71.74 | 77.97 | 0.65 | 0.70 | 0.65 | circle | 53 |
| 54 | Jones−UCL | 74.22 | 68.32 | 73.55 | 0.61 | 0.60 | 0.49 | 74.22 | 68.32 | 73.55 | 0.52 | 0.48 | 0.38 | Jones−UCL | 54 |
| 55 | HHpred2 | 74.07 | 68.69 | 75.65 | 0.60 | 0.62 | 0.61 | 74.07 | 68.69 | 75.65 | 0.51 | 0.50 | 0.51 | HHpred2 | 55 |
| 56 | DBAKER | 74.07 | 67.96 | 77.50 | 0.60 | 0.58 | 0.71 | 75.47 | 71.22 | 81.50 | 0.60 | 0.67 | 0.87 | DBAKER | 56 |
| 57 | 3DShot2 | 74.07 | 67.86 | 77.09 | 0.60 | 0.58 | 0.69 | 74.07 | 67.86 | 77.09 | 0.51 | 0.45 | 0.60 | 3DShot2 | 57 |
| 58 | SAMUDRALA | 74.07 | 67.65 | 78.05 | 0.60 | 0.56 | 0.74 | 75.47 | 70.34 | 80.15 | 0.60 | 0.61 | 0.78 | SAMUDRALA | 58 |
| 59 | fais−server | 73.91 | 69.56 | 74.57 | 0.59 | 0.67 | 0.55 | 73.91 | 69.56 | 74.57 | 0.50 | 0.56 | 0.45 | fais−server | 59 |
| 60 | ricardo | 73.60 | 66.46 | 73.63 | 0.57 | 0.49 | 0.50 | 74.22 | 67.70 | 73.63 | 0.52 | 0.44 | 0.39 | ricardo | 60 |
| 61 | A−TASSER | 73.60 | 66.46 | 75.17 | 0.57 | 0.49 | 0.58 | 73.91 | 68.84 | 75.17 | 0.50 | 0.51 | 0.48 | A−TASSER | 61 |
| 62 | BAKER−ROBETTA | 73.60 | 65.84 | 77.31 | 0.57 | 0.46 | 0.70 | 74.22 | 69.56 | 77.31 | 0.52 | 0.56 | 0.61 | BAKER−ROBETTA | 62 |
| 63 | PSI | 73.45 | 69.31 | 74.14 | 0.56 | 0.66 | 0.53 | 75.00 | 70.03 | 75.13 | 0.57 | 0.59 | 0.48 | PSI | 63 |
| 64 | Softberry | 72.36 | 62.22 | 70.39 | 0.50 | 0.25 | 0.32 | 72.36 | 62.22 | 70.39 | 0.40 | 0.09 | 0.19 | Softberry | 64 |
| 65 | nFOLD3 | 72.20 | 65.79 | 75.22 | 0.49 | 0.46 | 0.59 | 75.93 | 69.72 | 76.72 | 0.63 | 0.57 | 0.58 | nFOLD3 | 65 |
| 66 | AMU−Biology | 71.89 | 67.55 | 72.58 | 0.48 | 0.56 | 0.44 | 71.89 | 67.55 | 72.58 | 0.37 | 0.43 | 0.33 | AMU−Biology | 66 |
| 67 | SAM−T08−human | 71.89 | 66.41 | 76.46 | 0.48 | 0.49 | 0.65 | 74.84 | 69.93 | 76.46 | 0.56 | 0.58 | 0.56 | SAM−T08−human | 67 |
| 68 | SAM−T08−server | 71.43 | 65.32 | 74.56 | 0.45 | 0.43 | 0.55 | 72.98 | 67.18 | 75.75 | 0.44 | 0.41 | 0.52 | SAM−T08−server | 68 |
| 69 | SHORTLE | 70.34 | 63.09 | 69.97 | 0.39 | 0.30 | 0.30 | 70.34 | 63.09 | 69.97 | 0.28 | 0.14 | 0.17 | SHORTLE | 69 |
| 70 | PS2−server | 70.19 | 63.98 | 70.99 | 0.38 | 0.35 | 0.35 | 70.19 | 63.98 | 70.99 | 0.27 | 0.20 | 0.23 | PS2−server | 70 |
| 71 | PS2−manual | 70.19 | 63.98 | 70.99 | 0.38 | 0.35 | 0.35 | 70.19 | 63.98 | 70.99 | 0.27 | 0.20 | 0.23 | PS2−manual | 71 |
| 72 | SMEG−CCP | 70.19 | 60.87 | 69.69 | 0.38 | 0.17 | 0.28 | 70.19 | 60.87 | 69.69 | 0.27 | 0.15 | SMEG−CCP | 72 | |
| 73 | FALCON_CONSENSUS | 69.25 | 63.87 | 65.86 | 0.33 | 0.35 | 0.07 | 69.25 | 63.87 | 65.86 | 0.21 | 0.19 | FALCON_CONSENSUS | 73 | |
| 74 | FALCON | 69.25 | 63.87 | 65.86 | 0.33 | 0.35 | 0.07 | 69.25 | 63.87 | 65.86 | 0.21 | 0.19 | FALCON | 74 | |
| 75 | LevittGroup | 68.94 | 60.87 | 68.75 | 0.31 | 0.17 | 0.23 | 69.41 | 62.27 | 69.67 | 0.22 | 0.09 | 0.15 | LevittGroup | 75 |
| 76 | TJ_Jiang | 68.63 | 63.25 | 68.07 | 0.29 | 0.31 | 0.19 | 72.36 | 69.25 | 69.59 | 0.40 | 0.54 | 0.14 | TJ_Jiang | 76 |
| 77 | fais@hgc | 68.48 | 62.06 | 69.29 | 0.29 | 0.24 | 0.26 | 70.19 | 63.56 | 73.05 | 0.27 | 0.17 | 0.35 | fais@hgc | 77 |
| 78 | GeneSilicoMetaServer | 66.77 | 61.70 | 64.28 | 0.19 | 0.22 | 74.69 | 69.82 | 75.98 | 0.55 | 0.58 | 0.53 | GeneSilicoMetaServer | 78 | |
| 79 | SAM−T06−server | 66.30 | 59.06 | 67.20 | 0.16 | 0.07 | 0.15 | 71.89 | 66.92 | 74.19 | 0.37 | 0.39 | 0.42 | SAM−T06−server | 79 |
| 80 | pipe_int | 65.22 | 59.21 | 65.68 | 0.10 | 0.08 | 0.06 | 65.22 | 59.21 | 65.68 | pipe_int | 80 | |||
| 81 | FAMSD | 63.82 | 54.50 | 60.75 | 0.03 | 69.10 | 62.16 | 66.91 | 0.20 | 0.08 | FAMSD | 81 | |||
| 82 | Kolinski | 61.65 | 51.81 | 62.86 | 61.80 | 51.81 | 63.04 | Kolinski | 82 | ||||||
| 83 | FEIG | 54.81 | 43.94 | 56.13 | 74.53 | 69.36 | 76.35 | 0.54 | 0.55 | 0.55 | FEIG | 83 | |||
| 84 | Phyre_de_novo | 53.42 | 46.89 | 47.96 | 53.42 | 46.89 | 48.39 | Phyre_de_novo | 84 | ||||||
| 85 | FLOUDAS | 52.48 | 37.89 | 55.71 | 54.04 | 39.55 | 56.50 | FLOUDAS | 85 | ||||||
| 86 | Jiang_Zhu | 45.03 | 36.65 | 48.67 | 45.03 | 36.65 | 48.67 | Jiang_Zhu | 86 | ||||||
| 87 | LOOPP_Server | 43.79 | 35.20 | 37.09 | 47.98 | 38.15 | 39.99 | LOOPP_Server | 87 | ||||||
| 88 | huber−torda−server | 42.70 | 31.11 | 36.35 | 46.58 | 36.59 | 43.52 | huber−torda−server | 88 | ||||||
| 89 | FOLDpro | 42.55 | 36.75 | 37.13 | 75.62 | 68.89 | 75.51 | 0.61 | 0.52 | 0.50 | FOLDpro | 89 | |||
| 90 | Wolfson−FOBIA | 39.29 | 35.04 | 47.52 | 39.29 | 35.04 | 47.52 | Wolfson−FOBIA | 90 | ||||||
| 91 | Hao_Kihara | 38.66 | 36.70 | 36.18 | 39.75 | 38.09 | 36.89 | Hao_Kihara | 91 | ||||||
| 92 | BioSerf | 38.20 | 36.96 | 39.17 | 38.20 | 36.96 | 39.17 | BioSerf | 92 | ||||||
| 93 | Phyre2 | 38.04 | 33.18 | 44.79 | 39.60 | 37.53 | 45.42 | Phyre2 | 93 | ||||||
| 94 | Phragment | 37.73 | 32.87 | 46.18 | 38.51 | 37.06 | 46.18 | Phragment | 94 | ||||||
| 95 | mGenTHREADER | 37.58 | 36.13 | 32.80 | 37.58 | 36.13 | 32.80 | mGenTHREADER | 95 | ||||||
| 96 | fleil | 37.42 | 36.08 | 35.69 | 38.66 | 36.39 | 36.51 | fleil | 96 | ||||||
| 97 | Poing | 37.42 | 32.45 | 44.53 | 38.66 | 37.06 | 45.64 | Poing | 97 | ||||||
| 98 | CpHModels | 37.11 | 35.25 | 15.21 | 37.11 | 35.25 | 15.21 | CpHModels | 98 | ||||||
| 99 | 3D−JIGSAW_AEP | 37.11 | 35.15 | 36.53 | 38.82 | 36.13 | 37.03 | 3D−JIGSAW_AEP | 99 | ||||||
| 100 | panther_server | 36.80 | 33.59 | 15.50 | 37.58 | 36.13 | 19.01 | panther_server | 100 | ||||||
| 101 | 3D−JIGSAW_V3 | 34.94 | 32.35 | 32.86 | 38.20 | 36.23 | 40.09 | 3D−JIGSAW_V3 | 101 | ||||||
| 102 | GS−MetaServer2 | 32.14 | 29.24 | 31.15 | 74.69 | 69.82 | 75.98 | 0.55 | 0.58 | 0.53 | GS−MetaServer2 | 102 | |||
| 103 | SAM−T02−server | 31.37 | 27.85 | 16.34 | 31.37 | 28.88 | 24.94 | SAM−T02−server | 103 | ||||||
| 104 | Pushchino | 31.21 | 28.31 | 30.41 | 31.21 | 28.31 | 30.41 | Pushchino | 104 | ||||||
| 105 | xianmingpan | 28.73 | 25.62 | 34.40 | 28.73 | 25.62 | 34.40 | xianmingpan | 105 | ||||||
| 106 | Distill | 28.73 | 23.03 | 35.39 | 29.35 | 23.96 | 36.75 | Distill | 106 | ||||||
| 107 | ACOMPMOD | 28.42 | 24.95 | 32.69 | 46.27 | 39.29 | 46.07 | ACOMPMOD | 107 | ||||||
| 108 | FUGUE_KM | 27.02 | 24.84 | 26.76 | 45.65 | 37.68 | 42.89 | FUGUE_KM | 108 | ||||||
| 109 | PRI−Yang−KiharA | 26.71 | 23.60 | 33.38 | 26.71 | 23.60 | 33.38 | PRI−Yang−KiharA | 109 | ||||||
| 110 | StruPPi | 25.31 | 20.34 | 23.48 | 29.04 | 24.17 | 25.59 | StruPPi | 110 | ||||||
| 111 | DistillSN | 21.74 | 17.75 | 23.28 | 21.74 | 17.75 | 24.68 | DistillSN | 111 | ||||||
| 112 | MUFOLD−Server | 20.50 | 17.60 | 21.33 | 20.50 | 17.60 | 21.35 | MUFOLD−Server | 112 | ||||||
| 113 | MUFOLD−MD | 20.03 | 16.25 | 30.26 | 26.86 | 23.81 | 32.83 | MUFOLD−MD | 113 | ||||||
| 114 | SAINT1 | 19.88 | 16.67 | 28.97 | 19.88 | 16.67 | 28.97 | SAINT1 | 114 | ||||||
| 115 | Wolynes | 19.57 | 17.18 | 28.67 | 19.88 | 17.18 | 29.34 | Wolynes | 115 | ||||||
| 116 | keasar | 19.41 | 13.35 | 27.29 | 75.31 | 72.20 | 77.78 | 0.59 | 0.73 | 0.64 | keasar | 116 | |||
| 117 | MeilerLabRene | 18.79 | 17.29 | 26.01 | 19.10 | 17.29 | 29.44 | MeilerLabRene | 117 | ||||||
| 118 | RBO−Proteus | 17.70 | 14.29 | 29.12 | 20.19 | 16.36 | 29.12 | RBO−Proteus | 118 | ||||||
| 119 | rehtnap | 16.61 | 15.37 | 5.90 | 16.61 | 15.37 | 5.90 | rehtnap | 119 | ||||||
| 120 | Scheraga | 15.84 | 13.04 | 23.23 | 23.45 | 17.13 | 25.02 | Scheraga | 120 | ||||||
| 121 | schenk−torda−server | 14.91 | 12.42 | 19.08 | 14.91 | 12.42 | 19.08 | schenk−torda−server | 121 | ||||||
| 122 | RANDOM | 14.51 | 12.44 | 14.51 | 14.51 | 12.44 | 14.51 | RANDOM | 122 | ||||||
| 123 | mahmood−torda−server | 13.51 | 12.32 | 21.71 | 14.44 | 12.32 | 21.71 | mahmood−torda−server | 123 | ||||||
| 124 | DelCLab | 11.96 | 9.99 | 13.34 | 15.68 | 14.96 | 22.77 | DelCLab | 124 | ||||||
| 125 | TWPPLAB | 11.18 | 9.89 | 8.85 | 11.18 | 9.89 | 8.85 | TWPPLAB | 125 | ||||||
| 126 | OLGAFS | 11.03 | 9.37 | 3.20 | 15.22 | 12.63 | 9.35 | OLGAFS | 126 | ||||||
| 127 | mariner1 | 8.85 | 8.64 | 46.27 | 35.61 | 41.20 | mariner1 | 127 | |||||||
| 128 | Abagyan | Abagyan | 128 | ||||||||||||
| 129 | BHAGEERATH | BHAGEERATH | 129 | ||||||||||||
| 130 | CBSU | CBSU | 130 | ||||||||||||
| 131 | FEIG_REFINE | FEIG_REFINE | 131 | ||||||||||||
| 132 | Fiser−M4T | Fiser−M4T | 132 | ||||||||||||
| 133 | HCA | HCA | 133 | ||||||||||||
| 134 | Handl−Lovell | Handl−Lovell | 134 | ||||||||||||
| 135 | JIVE08 | JIVE08 | 135 | ||||||||||||
| 136 | LEE−SERVER | LEE−SERVER | 136 | ||||||||||||
| 137 | Linnolt−UH−CMB | Linnolt−UH−CMB | 137 | ||||||||||||
| 138 | NIM2 | NIM2 | 138 | ||||||||||||
| 139 | Nano_team | Nano_team | 139 | ||||||||||||
| 140 | NirBenTal | NirBenTal | 140 | ||||||||||||
| 141 | Ozkan−Shell | Ozkan−Shell | 141 | ||||||||||||
| 142 | PHAISTOS | PHAISTOS | 142 | ||||||||||||
| 143 | POISE | POISE | 143 | ||||||||||||
| 144 | PZ−UAM | PZ−UAM | 144 | ||||||||||||
| 145 | ProtAnG | ProtAnG | 145 | ||||||||||||
| 146 | ProteinShop | ProteinShop | 146 | ||||||||||||
| 147 | RPFM | RPFM | 147 | ||||||||||||
| 148 | Sasaki−Cetin−Sasai | Sasaki−Cetin−Sasai | 148 | ||||||||||||
| 149 | ShakAbInitio | ShakAbInitio | 149 | ||||||||||||
| 150 | TsaiLab | TsaiLab | 150 | ||||||||||||
| 151 | UCDavisGenome | UCDavisGenome | 151 | ||||||||||||
| 152 | YASARA | YASARA | 152 | ||||||||||||
| 153 | YASARARefine | YASARARefine | 153 | ||||||||||||
| 154 | Zhou−SPARKS | Zhou−SPARKS | 154 | ||||||||||||
| 155 | dill_ucsf | dill_ucsf | 155 | ||||||||||||
| 156 | dill_ucsf_extended | dill_ucsf_extended | 156 | ||||||||||||
| 157 | igor | igor | 157 | ||||||||||||
| 158 | jacobson | jacobson | 158 | ||||||||||||
| 159 | mti | mti | 159 | ||||||||||||
| 160 | mumssp | mumssp | 160 | ||||||||||||
| 161 | psiphifoldings | psiphifoldings | 161 | ||||||||||||
| 162 | rivilo | rivilo | 162 | ||||||||||||
| 163 | sessions | sessions | 163 | ||||||||||||
| 164 | taylor | taylor | 164 | ||||||||||||
| 165 | test_http_server_01 | test_http_server_01 | 165 | ||||||||||||
| 166 | tripos_08 | tripos_08 | 166 | ||||||||||||