T0478
conserved metalloprotein from Bacillus cereus
Target sequence:
>T0478 Hypothetical protein, Bacillus cereus, 283 residues
MSLDTGVTSVMFVERSLNEIRFWSRIMKEHSFFLRLGFRCEDTQLIEEANQFYRLFEHIEQIAHSYTNETDPEQIKRFNAEVQQAATNIWGFKRKILGLILTCKLPGQNNFPLLVDHTSREADYFRKRLIQLNEGKLDALPDAIIKENVFFLRIMADHAKFIGHLLDPSERKLVDTARNFSNDFDELMYQAIDLESMKPQSQTAPLLDQFLDQNRVSVASLRDFKKTARDLIEQCKIKSIIHPLLADHVFREADRFLEIIDMYDVHLTNIQSQPREGHHHHHH
Structure:
Determined by:
NYSGRC
PDB ID: 3d19
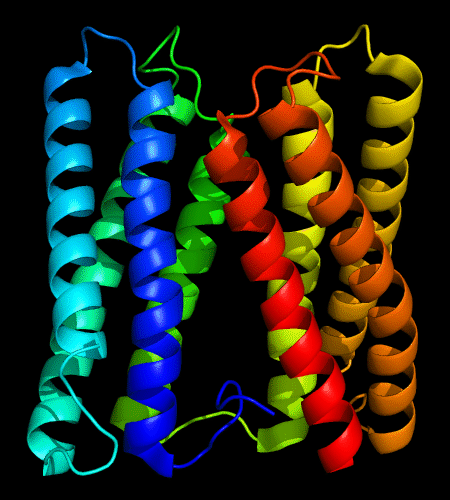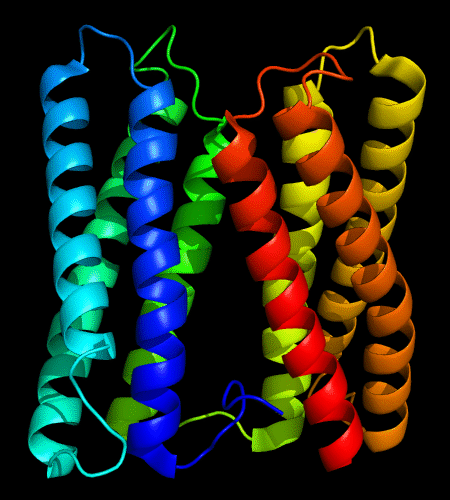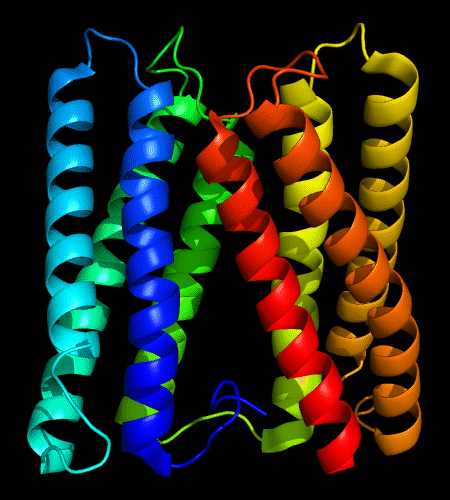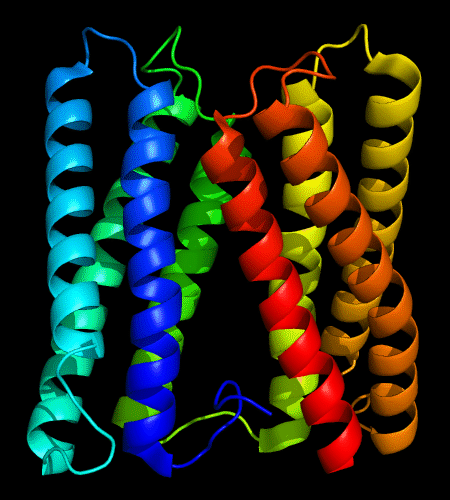
Cartoon diagram of 478: 3d19 chain A
Domains: PyMOL of domains
Two domains, duplication. Residue ranges in PDB: 11-144 and 145-274. Residue ranges in target: 4-137 and 138-267.
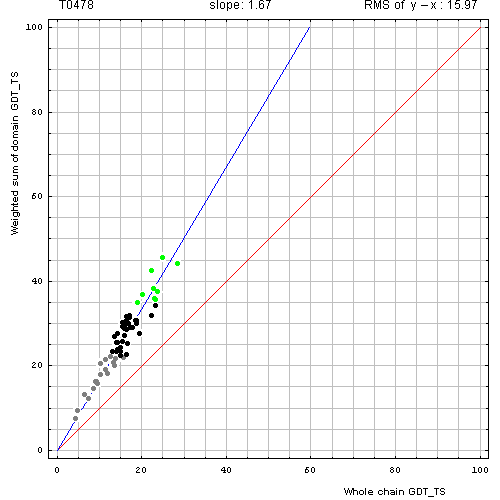
Correlation between weighted by the number of residues sum of GDT-TS scores for domain-based evlatuation (y, vertical axis)
and whole chain GDT-TS (x, horizontal axis).
To compute the weighted sum, GDT-TS for each domain was multiplied by the domain length, and this sum was divided by the sum of domain lengths. Each point represents first server model. Green, gray and black points are top 10, bottom 25% and the rest of prediction models. Blue line is the best-fit slope line (intersection 0) to the top 10 server models. Red line is the diagonal. Slope and root mean square y-x distance for the top 10 models (average difference between the weighted sum of domain GDT-TS scores and the whole chain GDT-TS score) are shown above the plot.
Structure classification:
Each domain is a 4-helical bundle starting with a left-handed triplet of α-helices, similarly to Bromodomain-like fold. Structure of each domain matched closely 4 long helices in RNA-binding protein She2p fold with the fifth helix removed.
CASP category:
Whole chain: Free modeling.
1st domain: Fold recognition. 2nd domain: Fold recognition.
Closest templates:
None found by the servers, although many bundles with 3 helices offer a good start. Reasonable templates for each domain could be up-and-down 4-helical bundles with the left-handed arrangement of the first 3 helices, e.g. 2e0a. This target should not be in the "server only" category, as it offers very challenging prediction opportunities.
Target sequence - PDB file inconsistencies:
T0478 3d19.pdb T0478.pdb PyMOL PyMOL of domains
T0478 1 MSLDTGVTSVMFVERSLNEIRFWSRIMKEHSFFLRLGFRCEDTQLIEEANQFYRLFEHIEQIAHSYTNETDPEQIKRFNAEVQQAATNIWGFKRKILGLILTCKLPGQNNFPLLVDHTSREADYFRKRLIQLNEGKLDALPDAIIKENVFFLRIMADHAKFIGHLLDPSERKLVDTARNFSNDFDELMYQAIDLESMKPQSQTAPLLDQFLDQNRVSVASLRDFKKTARDLIEQCKIKSIIHPLLADHVFREADRFLEIIDMYDVHLTNIQSQPREGHHHHHH 283 ~~~||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~~~~~~~~~~~~~~ 3d19A 11 ---DTGVTSVMFVERSLNEIRFWSRIMKEHSFFLRLGFRCEDTQLIEEANQFYRLFEHIEQIAHSYTNETDPEQIKRFNAEVQQAATNIWGFKRKILGLILTCKLPGQNNFPLLVDHTSREADYFRKRLIQLNEGKLDALPDAIIKENVFFLRIMADHAKFIGHLLDPSERKLVDTARNFSNDFDELMYQAIDLESMKPQSQTAPLLDQFLDQNRVSVASLRDFKKTARDLIEQCKIKSIIHPLLADHVFREADRFLEIIDMYDVHL---------------- 274
2-domain protein, both domains used in evaluation:
1st domain: target 4-137 ; pdb 11-144
2nd domain: target 138-267 ; pdb 145-274
Sequence classification:
Server predictions:
T0478:pdb 11-274:seq 4-267:FM; alignment
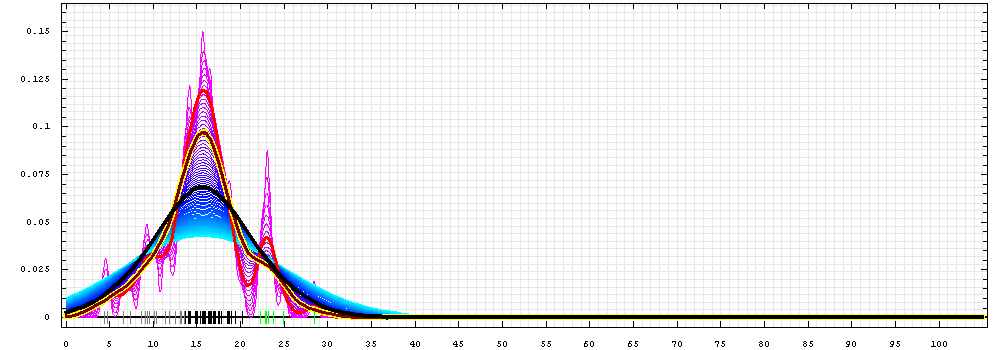
First models for T0478:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
478_1:pdb 11-144:seq 4-137:FR; alignment
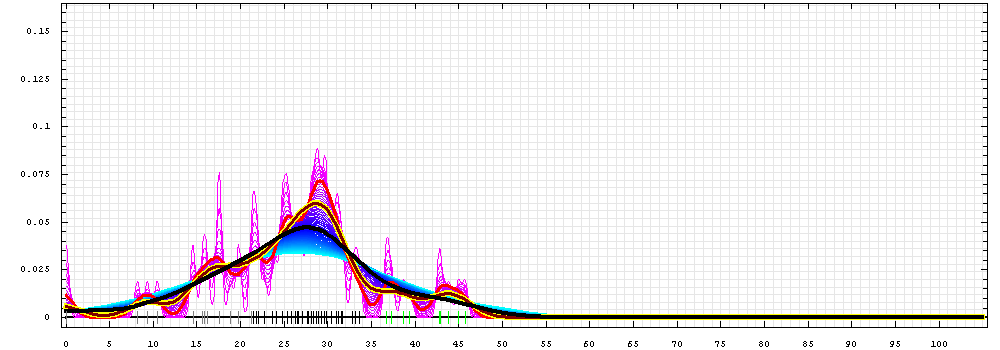
First models for T0478_1:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
478_2:pdb 145-274:seq 138-267:FR; alignment
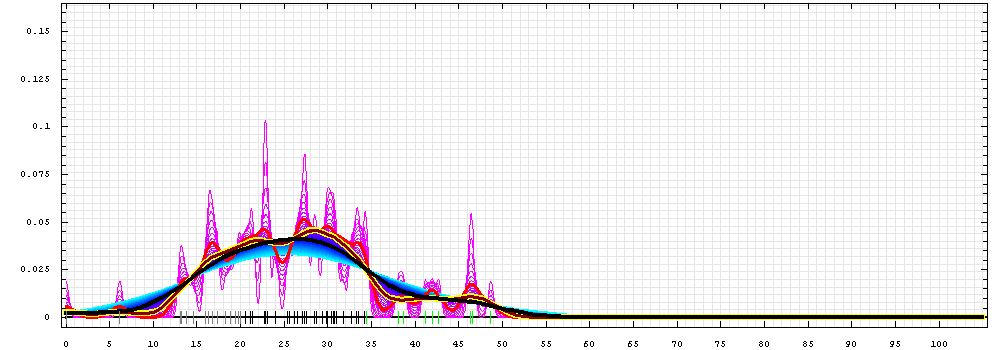
First models for T0478_2:
Gaussian kernel density estimation
for GDT-TS scores of the
first server models, plotted at various bandwidths (=standard deviations).
The GDT-TS scores are shown as a spectrum along
the horizontal axis: each bar represents first server model. The bars are
colored
green, gray and black for top 10, bottom 25% and the rest of servers.
The family of curves with varying
bandwidth is shown. Bandwidth varies from 0.3 to 8.2 GDT-TS % units
with a step of 0.1, which corresponds to the
color ramp from magenta through blue to cyan. Thicker curves: red,
yellow-framed brown and black, correspond to bandwidths 1, 2 and 4
respectively.
click on a score in the table below to display the model in PyMOL
| # | GROUP ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | TS ↓ | TR ↓ | CS ↓ | ↓ GROUP | # |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T0478 | 478_1 | 478_2 | T0478 | 478_1 | 478_2 | T0478 | 478_1 | 478_2 | T0478 | 478_1 | 478_2 | ||||||||||||||||||||||||||||
| First score | First Z-score | Best score | Best Z-score | ||||||||||||||||||||||||||||||||||||
| 1 | pro−sp3−TASSER | 28.41 | 19.25 | 34.21 | 45.71 | 25.43 | 44.16 | 42.69 | 31.60 | 40.23 | 3.33 | 2.31 | 1.55 | 2.54 | 0.70 | 1.49 | 1.91 | 1.57 | 1.17 | 28.41 | 19.25 | 34.21 | 49.44 | 33.15 | 44.49 | 42.69 | 34.42 | 40.23 | 3.08 | 1.64 | 1.52 | 2.70 | 1.62 | 1.37 | 1.51 | 1.50 | 1.10 | pro−sp3−TASSER | 1 |
| 2 | FALCON | 24.91 | 21.02 | 33.63 | 42.72 | 24.75 | 43.91 | 48.65 | 39.55 | 40.79 | 2.35 | 2.97 | 1.51 | 2.11 | 0.56 | 1.47 | 2.67 | 2.95 | 1.21 | 24.91 | 21.02 | 33.63 | 45.90 | 34.95 | 43.91 | 48.65 | 39.55 | 40.79 | 2.06 | 2.27 | 1.47 | 2.17 | 2.00 | 1.32 | 2.31 | 2.41 | 1.15 | FALCON | 2 |
| 3 | FALCON_CONSENSUS | 23.77 | 14.17 | 28.42 | 33.21 | 32.09 | 40.12 | 41.92 | 18.91 | 34.75 | 2.03 | 0.42 | 1.15 | 0.77 | 2.07 | 1.19 | 1.81 | 0.84 | 24.91 | 21.02 | 33.63 | 42.72 | 32.09 | 43.91 | 48.65 | 39.55 | 40.79 | 2.06 | 2.27 | 1.47 | 1.70 | 1.39 | 1.32 | 2.31 | 2.41 | 1.15 | FALCON_CONSENSUS | 3 | |
| 4 | RBO−Proteus | 23.20 | 19.60 | 26.05 | 44.96 | 37.87 | 42.53 | 23.08 | 22.56 | 27.67 | 1.87 | 2.44 | 0.98 | 2.43 | 3.25 | 1.36 | 0.41 | 23.67 | 20.33 | 28.98 | 45.71 | 39.12 | 45.52 | 26.35 | 24.61 | 30.11 | 1.69 | 2.03 | 1.07 | 2.15 | 2.89 | 1.47 | 0.21 | RBO−Proteus | 4 | ||||
| 5 | circle | 23.20 | 17.93 | 3.66 | 18.84 | 17.72 | 46.54 | 35.83 | 35.38 | 1.87 | 1.82 | 2.40 | 2.31 | 0.88 | 23.20 | 17.93 | 13.41 | 30.97 | 27.24 | 18.56 | 46.54 | 35.83 | 35.38 | 1.56 | 1.16 | 0.36 | 2.02 | 1.75 | 0.67 | circle | 5 | ||||||||
| 6 | Pcons_dot_net | 23.01 | 19.03 | 5.25 | 19.78 | 18.53 | 46.35 | 38.27 | 38.03 | 1.82 | 2.23 | 2.38 | 2.73 | 1.04 | 23.30 | 19.13 | 8.17 | 21.08 | 20.21 | 0.58 | 46.73 | 38.27 | 38.44 | 1.59 | 1.59 | 2.05 | 2.19 | 0.94 | Pcons_dot_net | 6 | |||||||||
| 7 | Pcons_multi | 23.01 | 19.03 | 5.25 | 19.78 | 18.53 | 46.35 | 38.27 | 38.03 | 1.82 | 2.23 | 2.38 | 2.73 | 1.04 | 23.11 | 19.03 | 12.94 | 26.31 | 18.97 | 14.91 | 46.54 | 38.27 | 39.10 | 1.53 | 1.56 | 2.02 | 2.19 | 1.00 | Pcons_multi | 7 | |||||||||
| 8 | Zhang−Server | 22.82 | 16.00 | 39.28 | 38.62 | 33.40 | 47.94 | 38.08 | 26.80 | 46.72 | 1.76 | 1.10 | 1.90 | 1.53 | 2.34 | 1.77 | 1.32 | 0.74 | 1.57 | 28.41 | 23.74 | 44.27 | 49.63 | 39.05 | 52.39 | 44.42 | 33.59 | 51.73 | 3.08 | 3.25 | 2.38 | 2.73 | 2.88 | 2.10 | 1.74 | 1.36 | 2.10 | Zhang−Server | 8 |
| 9 | FAMSD | 22.25 | 18.09 | 17.29 | 42.91 | 34.83 | 37.60 | 20.58 | 18.53 | 16.58 | 1.60 | 1.88 | 0.37 | 2.14 | 2.63 | 1.00 | 22.25 | 18.09 | 17.29 | 42.91 | 34.83 | 37.60 | 30.00 | 25.00 | 29.51 | 1.28 | 1.22 | 0.06 | 1.73 | 1.98 | 0.73 | 0.16 | FAMSD | 9 | |||||
| 10 | MUFOLD−Server | 22.25 | 11.14 | 29.14 | 43.84 | 21.95 | 40.12 | 41.15 | 19.55 | 36.18 | 1.60 | 1.20 | 2.27 | 1.19 | 1.71 | 0.93 | 22.63 | 14.80 | 29.14 | 43.84 | 25.31 | 42.03 | 41.15 | 23.33 | 38.44 | 1.39 | 0.03 | 1.08 | 1.87 | 1.14 | 1.31 | 0.94 | MUFOLD−Server | 10 | |||||
| 11 | SAM−T08−server | 20.17 | 10.83 | 26.72 | 39.37 | 20.96 | 35.71 | 34.23 | 29.81 | 35.60 | 1.02 | 1.03 | 1.64 | 0.86 | 0.83 | 1.26 | 0.89 | 20.17 | 14.11 | 33.29 | 39.37 | 27.61 | 46.03 | 34.23 | 29.81 | 37.74 | 0.67 | 1.44 | 1.20 | 0.44 | 1.51 | 0.38 | 0.68 | 0.88 | SAM−T08−server | 11 | |||
| 12 | SAM−T06−server | 19.41 | 15.85 | 19.90 | 21.27 | 20.27 | 28.54 | 34.23 | 28.59 | 30.95 | 0.81 | 1.04 | 0.55 | 0.32 | 0.83 | 1.05 | 0.61 | 20.55 | 18.66 | 19.90 | 38.81 | 24.25 | 28.54 | 39.04 | 35.19 | 30.95 | 0.78 | 1.42 | 0.29 | 1.12 | 1.03 | 1.64 | 0.29 | SAM−T06−server | 12 | ||||
| 13 | fais−server | 18.94 | 9.38 | 33.66 | 31.53 | 25.56 | 43.81 | 38.65 | 19.61 | 40.90 | 0.68 | 1.51 | 0.53 | 0.73 | 1.46 | 1.39 | 1.22 | 19.22 | 14.62 | 33.66 | 37.87 | 27.24 | 43.81 | 38.65 | 27.18 | 40.90 | 0.40 | 1.47 | 0.98 | 0.36 | 1.31 | 0.97 | 0.22 | 1.16 | fais−server | 13 | |||
| 14 | MUFOLD−MD | 18.75 | 13.26 | 25.22 | 37.31 | 25.75 | 39.60 | 24.04 | 22.31 | 29.58 | 0.63 | 0.08 | 0.92 | 1.35 | 0.76 | 1.15 | 0.52 | 18.75 | 17.17 | 27.03 | 37.31 | 31.34 | 39.68 | 29.23 | 23.78 | 34.77 | 0.26 | 0.89 | 0.90 | 0.90 | 1.23 | 0.93 | 0.62 | MUFOLD−MD | 14 | ||||
| 15 | GS−MetaServer2 | 18.66 | 13.10 | 36.75 | 25.81 | 10.11 | 22.89 | 20.96 | 0.38 | 0.60 | 0.02 | 1.27 | 0.78 | 18.66 | 14.43 | 10.70 | 36.75 | 26.74 | 29.50 | 32.50 | 26.80 | 26.77 | 0.23 | 0.81 | 0.25 | 0.15 | 0.15 | GS−MetaServer2 | 15 | ||||||||||
| 16 | GeneSilicoMetaServer | 18.66 | 13.10 | 36.75 | 25.81 | 10.11 | 22.89 | 20.96 | 0.38 | 0.60 | 0.02 | 1.27 | 0.78 | 18.66 | 14.43 | 10.70 | 36.75 | 26.74 | 29.50 | 32.50 | 26.80 | 26.77 | 0.23 | 0.81 | 0.25 | 0.15 | 0.15 | GeneSilicoMetaServer | 16 | ||||||||||
| 17 | LEE−SERVER | 18.47 | 16.32 | 26.95 | 30.41 | 24.19 | 37.57 | 30.96 | 26.15 | 34.48 | 0.55 | 1.22 | 1.04 | 0.37 | 0.44 | 1.00 | 0.41 | 0.62 | 0.82 | 21.12 | 20.04 | 26.95 | 36.01 | 33.77 | 37.57 | 34.81 | 26.15 | 34.48 | 0.95 | 1.92 | 0.89 | 0.70 | 1.75 | 0.73 | 0.46 | 0.04 | 0.60 | LEE−SERVER | 17 |
| 18 | MULTICOM−CMFR | 17.80 | 12.40 | 20.74 | 23.69 | 20.59 | 24.39 | 34.42 | 25.19 | 36.95 | 0.36 | 0.61 | 0.02 | 0.85 | 0.46 | 0.97 | 18.09 | 14.87 | 20.74 | 30.97 | 26.62 | 32.77 | 34.42 | 29.55 | 36.95 | 0.07 | 0.06 | 0.36 | 0.23 | 0.29 | 0.41 | 0.64 | 0.81 | MULTICOM−CMFR | 18 | ||||
| 19 | MULTICOM−RANK | 17.61 | 13.04 | 21.43 | 24.81 | 21.58 | 25.94 | 33.27 | 21.99 | 36.24 | 0.31 | 0.66 | 0.13 | 0.70 | 0.93 | 17.90 | 15.31 | 21.43 | 28.17 | 27.80 | 28.15 | 33.27 | 29.30 | 36.24 | 0.01 | 0.22 | 0.42 | 0.48 | 0.26 | 0.59 | 0.75 | MULTICOM−RANK | 19 | ||||||
| 20 | MULTICOM−CLUSTER | 17.61 | 12.78 | 21.85 | 24.81 | 21.45 | 26.23 | 33.46 | 29.68 | 36.88 | 0.31 | 0.69 | 0.15 | 0.73 | 1.24 | 0.97 | 17.61 | 14.80 | 21.85 | 28.55 | 25.93 | 28.47 | 33.46 | 29.68 | 36.88 | 0.03 | 0.45 | 0.08 | 0.28 | 0.66 | 0.80 | MULTICOM−CLUSTER | 20 | ||||||
| 21 | MULTICOM−REFINE | 17.61 | 12.78 | 21.85 | 24.81 | 21.45 | 26.23 | 33.46 | 29.68 | 36.88 | 0.31 | 0.69 | 0.15 | 0.73 | 1.24 | 0.97 | 17.61 | 14.71 | 21.85 | 30.22 | 25.25 | 35.29 | 33.46 | 29.68 | 36.88 | 0.00 | 0.45 | 0.52 | 0.28 | 0.66 | 0.80 | MULTICOM−REFINE | 21 | ||||||
| 22 | PSI | 17.42 | 13.26 | 28.33 | 29.66 | 26.93 | 38.88 | 28.46 | 21.54 | 35.85 | 0.25 | 0.08 | 1.14 | 0.26 | 1.01 | 1.09 | 0.09 | 0.91 | 18.94 | 16.45 | 30.54 | 36.38 | 32.40 | 40.68 | 35.96 | 29.94 | 39.69 | 0.32 | 0.63 | 1.20 | 0.76 | 1.46 | 1.02 | 0.62 | 0.71 | 1.05 | PSI | 22 | |
| 23 | PS2−server | 17.14 | 15.37 | 24.83 | 32.84 | 29.35 | 31.82 | 29.81 | 29.42 | 36.60 | 0.18 | 0.86 | 0.90 | 0.71 | 1.50 | 0.57 | 0.26 | 1.19 | 0.95 | 17.14 | 15.37 | 24.83 | 32.84 | 29.35 | 42.16 | 30.58 | 29.42 | 36.60 | 0.24 | 0.71 | 0.23 | 0.81 | 1.16 | 0.62 | 0.78 | PS2−server | 23 | ||
| 24 | BAKER−ROBETTA | 17.05 | 15.72 | 28.28 | 33.58 | 30.54 | 41.48 | 30.00 | 23.27 | 33.01 | 0.15 | 0.99 | 1.14 | 0.82 | 1.75 | 1.29 | 0.28 | 0.12 | 0.73 | 17.23 | 16.04 | 28.28 | 33.58 | 30.54 | 41.48 | 32.69 | 28.08 | 35.40 | 0.48 | 1.01 | 0.34 | 1.06 | 1.09 | 0.18 | 0.38 | 0.68 | BAKER−ROBETTA | 24 | |
| 25 | MUProt | 16.86 | 15.21 | 17.47 | 28.36 | 20.02 | 28.53 | 31.73 | 29.42 | 26.19 | 0.10 | 0.80 | 0.38 | 0.08 | 0.32 | 0.50 | 1.19 | 0.31 | 17.80 | 15.91 | 21.85 | 30.22 | 25.00 | 35.16 | 33.85 | 30.19 | 36.94 | 0.43 | 0.45 | 0.51 | 0.33 | 0.75 | 0.81 | MUProt | 25 | ||||
| 26 | 3Dpro | 16.67 | 12.28 | 18.87 | 31.72 | 20.27 | 29.43 | 28.65 | 25.06 | 28.08 | 0.05 | 0.48 | 0.56 | 0.39 | 0.11 | 0.43 | 0.43 | 16.67 | 12.28 | 18.87 | 31.72 | 20.27 | 29.43 | 28.65 | 25.06 | 28.08 | 0.20 | 0.06 | 0.04 | 3Dpro | 26 | ||||||||
| 27 | MUSTER | 16.57 | 14.74 | 19.59 | 27.80 | 14.86 | 32.14 | 22.69 | 19.68 | 26.52 | 0.02 | 0.63 | 0.53 | 0.00 | 0.59 | 0.33 | 16.67 | 14.96 | 19.59 | 28.73 | 25.12 | 32.14 | 29.23 | 23.59 | 26.56 | 0.09 | 0.26 | 0.23 | MUSTER | 27 | |||||||||
| 28 | GS−KudlatyPred | 16.48 | 14.08 | 26.42 | 28.92 | 22.70 | 36.19 | 32.69 | 28.08 | 35.32 | 0.38 | 1.01 | 0.16 | 0.14 | 0.89 | 0.63 | 0.96 | 0.87 | 23.11 | 18.88 | 27.97 | 29.66 | 24.88 | 38.86 | 46.35 | 37.76 | 36.72 | 1.53 | 1.50 | 0.98 | 0.85 | 2.00 | 2.09 | 0.79 | GS−KudlatyPred | 28 | |||
| 29 | Phyre_de_novo | 16.48 | 13.70 | 16.32 | 28.92 | 25.31 | 36.19 | 16.35 | 14.23 | 15.79 | 0.24 | 0.30 | 0.16 | 0.67 | 0.89 | 16.48 | 13.70 | 17.25 | 32.09 | 26.37 | 38.32 | 22.50 | 21.67 | 22.33 | 0.06 | 0.12 | 0.17 | 0.80 | Phyre_de_novo | 29 | |||||||||
| 30 | keasar−server | 16.38 | 11.74 | 10.77 | 30.41 | 26.74 | 18.29 | 32.69 | 23.27 | 24.54 | 0.37 | 0.97 | 0.63 | 0.12 | 0.21 | 18.56 | 13.64 | 14.33 | 36.57 | 26.87 | 24.96 | 33.27 | 28.08 | 24.87 | 0.20 | 0.79 | 0.28 | 0.26 | 0.38 | keasar−server | 30 | ||||||||
| 31 | Poing | 16.29 | 14.49 | 19.94 | 26.49 | 21.52 | 29.70 | 30.77 | 26.92 | 29.94 | 0.54 | 0.56 | 0.41 | 0.38 | 0.76 | 0.54 | 16.38 | 14.49 | 23.09 | 27.43 | 23.32 | 32.92 | 32.31 | 26.92 | 33.45 | 0.56 | 0.30 | 0.13 | 0.17 | 0.51 | Poing | 31 | |||||||
| 32 | RANDOM | 16.03 | 13.39 | 16.03 | 25.81 | 22.41 | 25.81 | 28.41 | 23.55 | 28.41 | 0.12 | 0.28 | 0.08 | 0.12 | 0.08 | 0.17 | 0.45 | 16.03 | 13.39 | 16.03 | 25.81 | 22.41 | 25.81 | 28.41 | 23.55 | 28.41 | 0.07 | RANDOM | 32 | ||||||||||
| 33 | RAPTOR | 15.81 | 14.71 | 29.91 | 31.16 | 28.98 | 41.60 | 26.15 | 23.53 | 35.95 | 0.62 | 1.25 | 0.48 | 1.43 | 1.30 | 0.17 | 0.91 | 18.56 | 15.85 | 32.66 | 32.09 | 28.98 | 46.46 | 29.23 | 27.50 | 37.85 | 0.20 | 0.41 | 1.38 | 0.12 | 0.73 | 1.55 | 0.27 | 0.89 | RAPTOR | 33 | |||
| 34 | pipe_int | 15.81 | 14.08 | 21.81 | 30.97 | 27.49 | 31.55 | 29.42 | 24.30 | 31.37 | 0.38 | 0.69 | 0.45 | 1.12 | 0.55 | 0.21 | 0.30 | 0.63 | 15.81 | 14.08 | 21.81 | 30.97 | 27.49 | 31.55 | 29.42 | 24.30 | 31.37 | 0.45 | 0.41 | 0.17 | 0.32 | pipe_int | 34 | ||||||
| 35 | LOOPP_Server | 15.81 | 10.89 | 30.97 | 21.27 | 8.29 | 27.31 | 9.68 | 0.45 | 18.28 | 16.19 | 15.66 | 35.08 | 31.22 | 39.73 | 36.54 | 27.82 | 25.44 | 0.12 | 0.53 | 0.56 | 1.21 | 0.93 | 0.69 | 0.33 | LOOPP_Server | 35 | ||||||||||||
| 36 | CpHModels | 15.72 | 13.83 | 17.13 | 29.85 | 22.20 | 28.64 | 30.00 | 26.67 | 25.47 | 0.29 | 0.36 | 0.29 | 0.03 | 0.33 | 0.28 | 0.71 | 0.27 | 15.72 | 13.83 | 17.13 | 29.85 | 22.20 | 28.64 | 30.00 | 26.67 | 25.47 | 0.05 | 0.13 | CpHModels | 36 | ||||||||
| 37 | HHpred2 | 15.62 | 12.50 | 29.66 | 23.51 | 24.96 | 13.85 | 12.05 | 0.26 | 0.30 | 0.06 | 15.62 | 12.50 | 29.66 | 23.51 | 24.96 | 13.85 | 12.05 | HHpred2 | 37 | |||||||||||||||||||
| 38 | forecast | 15.44 | 13.07 | 13.83 | 28.17 | 22.33 | 26.81 | 30.39 | 24.10 | 21.41 | 0.00 | 0.13 | 0.05 | 0.06 | 0.20 | 0.33 | 0.27 | 0.02 | 16.57 | 14.93 | 17.66 | 29.29 | 23.88 | 29.74 | 30.39 | 24.10 | 26.39 | 0.08 | 0.09 | 0.00 | forecast | 38 | |||||||
| 39 | nFOLD3 | 15.44 | 12.72 | 24.37 | 29.66 | 17.85 | 30.55 | 30.58 | 25.06 | 37.26 | 0.86 | 0.26 | 0.47 | 0.36 | 0.43 | 0.99 | 17.80 | 13.86 | 24.37 | 29.66 | 23.13 | 32.78 | 36.35 | 27.37 | 37.26 | 0.67 | 0.29 | 0.67 | 0.25 | 0.84 | nFOLD3 | 39 | |||||||
| 40 | FFASsuboptimal | 15.44 | 10.48 | 0.30 | 27.61 | 15.05 | 24.84 | 22.69 | 21.03 | 0.05 | 15.72 | 12.31 | 5.05 | 30.97 | 25.43 | 26.80 | 28.08 | 21.09 | 23.93 | FFASsuboptimal | 40 | ||||||||||||||||||
| 41 | 3D−JIGSAW_V3 | 15.06 | 13.86 | 8.45 | 28.17 | 23.69 | 24.06 | 20.00 | 18.85 | 13.89 | 0.30 | 0.05 | 0.34 | 15.06 | 13.86 | 8.52 | 28.17 | 26.68 | 24.12 | 20.39 | 19.30 | 14.52 | 0.24 | 3D−JIGSAW_V3 | 41 | ||||||||||||||
| 42 | FFASstandard | 14.96 | 10.61 | 7.40 | 17.54 | 14.30 | 13.20 | 27.50 | 17.24 | 23.63 | 0.16 | 15.53 | 10.61 | 14.48 | 19.22 | 17.10 | 23.04 | 28.46 | 18.46 | 26.48 | FFASstandard | 42 | |||||||||||||||||
| 43 | FFASflextemplate | 14.96 | 10.61 | 7.40 | 17.54 | 14.30 | 13.20 | 27.50 | 17.24 | 23.63 | 0.16 | 15.53 | 10.61 | 14.48 | 19.22 | 17.10 | 23.04 | 28.46 | 18.46 | 26.48 | FFASflextemplate | 43 | |||||||||||||||||
| 44 | Phragment | 14.87 | 12.97 | 8.67 | 29.29 | 25.56 | 23.64 | 17.31 | 16.41 | 14.70 | 0.21 | 0.73 | 16.10 | 12.97 | 16.76 | 31.53 | 25.56 | 31.52 | 22.89 | 20.32 | 21.61 | 0.02 | 0.04 | 0.00 | 0.17 | Phragment | 44 | ||||||||||||
| 45 | COMA | 14.30 | 12.60 | 4.96 | 28.73 | 19.78 | 23.82 | 26.54 | 24.74 | 7.82 | 0.13 | 0.38 | 14.39 | 12.60 | 6.19 | 28.92 | 19.78 | 25.58 | 26.92 | 25.19 | 10.55 | COMA | 45 | ||||||||||||||||
| 46 | COMA−M | 14.30 | 12.60 | 4.96 | 28.73 | 19.78 | 23.82 | 26.54 | 24.74 | 7.82 | 0.13 | 0.38 | 14.39 | 12.60 | 6.19 | 28.92 | 19.78 | 25.58 | 26.92 | 25.19 | 10.55 | COMA−M | 46 | ||||||||||||||||
| 47 | Pcons_local | 14.30 | 12.53 | 27.05 | 26.68 | 11.09 | 24.04 | 23.65 | 0.96 | 0.19 | 18.09 | 12.53 | 8.17 | 27.05 | 26.68 | 11.09 | 36.73 | 25.13 | 38.05 | 0.07 | 0.24 | 0.72 | 0.91 | Pcons_local | 47 | ||||||||||||||
| 48 | BioSerf | 14.21 | 11.55 | 15.65 | 22.02 | 21.14 | 26.94 | 25.58 | 21.09 | 24.72 | 0.26 | 0.21 | 0.22 | 14.21 | 11.55 | 15.65 | 22.02 | 21.14 | 26.94 | 25.58 | 21.09 | 24.72 | BioSerf | 48 | |||||||||||||||
| 49 | 3D−JIGSAW_AEP | 14.11 | 12.37 | 6.08 | 26.68 | 23.45 | 19.48 | 19.81 | 17.50 | 14.25 | 0.29 | 14.30 | 13.04 | 6.15 | 27.24 | 26.12 | 20.13 | 19.81 | 18.78 | 14.49 | 0.12 | 3D−JIGSAW_AEP | 49 | ||||||||||||||||
| 50 | Distill | 14.02 | 13.89 | 28.17 | 25.37 | 25.12 | 38.61 | 25.39 | 25.39 | 36.19 | 0.31 | 1.13 | 0.63 | 1.07 | 0.49 | 0.93 | 14.30 | 14.11 | 28.41 | 25.75 | 25.37 | 39.31 | 26.15 | 25.39 | 36.19 | 1.02 | 0.89 | 0.74 | Distill | 50 | |||||||||
| 51 | mGenTHREADER | 13.73 | 12.25 | 7.26 | 24.07 | 19.96 | 18.52 | 19.42 | 16.86 | 17.77 | 13.73 | 12.25 | 7.26 | 24.07 | 19.96 | 18.52 | 19.42 | 16.86 | 17.77 | mGenTHREADER | 51 | ||||||||||||||||||
| 52 | Frankenstein | 13.64 | 12.44 | 16.57 | 26.31 | 22.57 | 30.05 | 27.31 | 24.23 | 23.30 | 0.32 | 0.11 | 0.44 | 0.29 | 0.14 | 17.52 | 14.55 | 16.57 | 31.34 | 27.11 | 30.05 | 34.81 | 26.35 | 23.30 | 0.01 | 0.33 | 0.03 | 0.46 | 0.07 | Frankenstein | 52 | ||||||||
| 53 | mariner1 | 13.54 | 10.64 | 22.76 | 18.28 | 3.23 | 17.31 | 14.17 | 16.86 | 14.96 | 11.80 | 29.10 | 25.12 | 28.86 | 28.46 | 26.54 | 17.34 | 0.09 | 0.10 | mariner1 | 53 | ||||||||||||||||||
| 54 | FOLDpro | 13.26 | 11.49 | 14.55 | 13.56 | 27.11 | 23.53 | 21.76 | 0.17 | 0.04 | 16.67 | 12.28 | 18.87 | 31.72 | 21.45 | 29.43 | 28.65 | 25.06 | 28.08 | 0.20 | 0.06 | 0.04 | FOLDpro | 54 | |||||||||||||||
| 55 | HHpred4 | 13.07 | 9.91 | 25.37 | 20.77 | 19.60 | 21.35 | 16.73 | 13.07 | 9.91 | 25.37 | 20.77 | 19.60 | 21.35 | 16.73 | HHpred4 | 55 | ||||||||||||||||||||||
| 56 | HHpred5 | 13.07 | 9.91 | 25.37 | 20.77 | 19.60 | 21.35 | 16.73 | 13.07 | 9.91 | 25.37 | 20.77 | 19.60 | 21.35 | 16.73 | HHpred5 | 56 | ||||||||||||||||||||||
| 57 | METATASSER | 12.50 | 9.91 | 16.82 | 21.45 | 18.04 | 30.78 | 22.89 | 15.83 | 22.75 | 0.34 | 0.49 | 0.10 | 21.50 | 17.17 | 33.78 | 37.31 | 29.17 | 42.07 | 32.88 | 27.56 | 41.82 | 1.06 | 0.89 | 1.48 | 0.90 | 0.77 | 1.15 | 0.21 | 0.29 | 1.24 | METATASSER | 57 | ||||||
| 58 | FEIG | 11.84 | 9.19 | 2.29 | 17.54 | 15.05 | 17.28 | 18.85 | 15.51 | 9.65 | 15.06 | 13.04 | 10.95 | 21.08 | 17.60 | 28.43 | 19.04 | 18.08 | 14.33 | FEIG | 58 | ||||||||||||||||||
| 59 | 3DShot2 | 11.36 | 11.27 | 0.62 | 21.83 | 21.45 | 3.67 | 21.15 | 20.96 | 20.82 | 11.36 | 11.27 | 0.62 | 21.83 | 21.45 | 3.67 | 21.15 | 20.96 | 20.82 | 3DShot2 | 59 | ||||||||||||||||||
| 60 | Phyre2 | 11.36 | 10.04 | 3.88 | 21.45 | 19.59 | 20.25 | 16.73 | 15.32 | 9.08 | 14.02 | 12.25 | 3.88 | 23.69 | 19.71 | 24.33 | 23.85 | 19.49 | 9.08 | Phyre2 | 60 | ||||||||||||||||||
| 61 | schenk−torda−server | 10.32 | 8.30 | 17.54 | 15.05 | 3.50 | 18.27 | 14.74 | 1.20 | 10.32 | 8.30 | 17.91 | 15.05 | 3.50 | 18.27 | 14.74 | 4.26 | schenk−torda−server | 61 | ||||||||||||||||||||
| 62 | FUGUE_KM | 10.13 | 9.41 | 0.00 | 0.00 | 20.58 | 19.10 | 18.47 | 13.67 | 1.65 | 36.38 | 26.93 | 10.90 | 33.08 | 23.46 | 25.53 | 0.18 | 0.76 | 0.29 | 0.23 | FUGUE_KM | 62 | |||||||||||||||||
| 63 | Pushchino | 9.56 | 7.10 | 15.86 | 11.01 | 15.96 | 15.19 | 9.56 | 7.10 | 15.86 | 11.01 | 15.96 | 15.19 | Pushchino | 63 | ||||||||||||||||||||||||
| 64 | mahmood−torda−server | 9.28 | 8.21 | 15.67 | 13.06 | 10.56 | 16.73 | 13.91 | 4.39 | 10.89 | 8.55 | 19.03 | 14.80 | 10.56 | 19.61 | 14.04 | 9.80 | mahmood−torda−server | 64 | ||||||||||||||||||||
| 65 | huber−torda−server | 9.09 | 8.14 | 16.23 | 14.30 | 16.35 | 15.83 | 10.98 | 9.69 | 18.28 | 16.91 | 1.43 | 19.42 | 17.11 | huber−torda−server | 65 | |||||||||||||||||||||||
| 66 | OLGAFS | 8.62 | 7.17 | 14.55 | 12.69 | 14.62 | 13.46 | 12.60 | 8.49 | 16.23 | 14.24 | 19.23 | 18.20 | OLGAFS | 66 | ||||||||||||||||||||||||
| 67 | SAM−T02−server | 7.39 | 5.93 | 10.45 | 8.09 | 13.08 | 10.96 | 10.89 | 9.75 | 21.45 | 19.22 | 17.69 | 16.73 | SAM−T02−server | 67 | ||||||||||||||||||||||||
| 68 | ACOMPMOD | 6.53 | 5.27 | 0.00 | 0.00 | 13.27 | 10.71 | 15.72 | 12.66 | 2.82 | 25.19 | 24.94 | 4.47 | 32.12 | 26.15 | 27.00 | 0.10 | 0.04 | ACOMPMOD | 68 | |||||||||||||||||||
| 69 | rehtnap | 4.74 | 4.74 | 9.33 | 9.33 | 0.00 | 0.00 | 4.74 | 4.74 | 9.33 | 9.33 | 0.00 | 0.00 | rehtnap | 69 | ||||||||||||||||||||||||
| 70 | panther_server | 4.36 | 3.54 | 8.21 | 6.59 | 6.15 | 5.26 | 4.36 | 3.54 | 8.21 | 6.59 | 6.15 | 5.26 | panther_server | 70 | ||||||||||||||||||||||||
| 71 | BHAGEERATH | 0.12 | 0.28 | 0.08 | 0.12 | 0.08 | 0.17 | 0.45 | 0.07 | BHAGEERATH | 71 | ||||||||||||||||||||||||||||
| 72 | Fiser−M4T | 0.12 | 0.28 | 0.08 | 0.12 | 0.08 | 0.17 | 0.45 | 0.07 | Fiser−M4T | 72 | ||||||||||||||||||||||||||||
| 73 | YASARA | 0.12 | 0.28 | 0.08 | 0.12 | 0.08 | 0.17 | 0.45 | 0.07 | YASARA | 73 | ||||||||||||||||||||||||||||
| 74 | test_http_server_01 | 0.12 | 0.28 | 0.08 | 0.12 | 0.08 | 0.17 | 0.45 | 0.07 | test_http_server_01 | 74 | ||||||||||||||||||||||||||||