Where are the new folds?
Were did the new folds go? It was a prominent category in early CASPs. Now, it is not even possible to define it as a "category". New folds are approaching extinction, and fast. In fact, it is likely that the majority of structure types are already known for water soluble proteins. We agree with the seminal paper from Jeff Skolnick group 1) that the structure space knowledge is close to complete as far as distinct types of secondary structure packing are concerned. HOWEVER, this is very far from saying that we know how to map sequence space on structure space, as deduction of many folds from sequence is currently not possible, and CASP8 results show it. Many families with "old" fold are not predictable from sequence. For instance, no server found the correct template for T0460, and confident predictions were not possible for T0465, T0466, etc. Thus, structural biologists still have a long road ahead as structural genomics reveals and will continue to find many wonderful examples of unusual occurrences of such sequence-unpredictable "old" folds hiding among the semi-random families being structurally characterized.
Back to new folds, i.e. distinct cores of secondary structural elements with connections and spatial arrangement not observed before. Although fold definition is subject to debate, experts frequently agree on what looks like a "new fold". Several experts in our group inspected CASP8 target structures and concluded that only two domains represent new folds. These were T0397_1 and T0496_1:
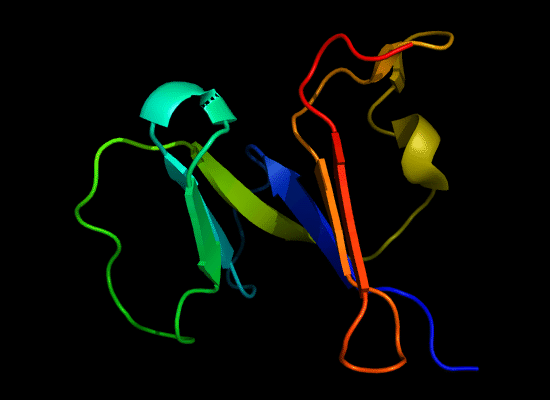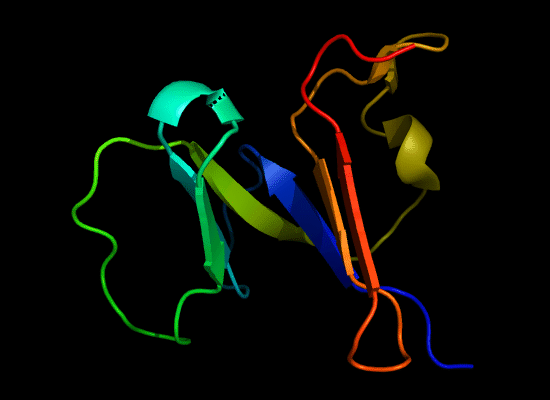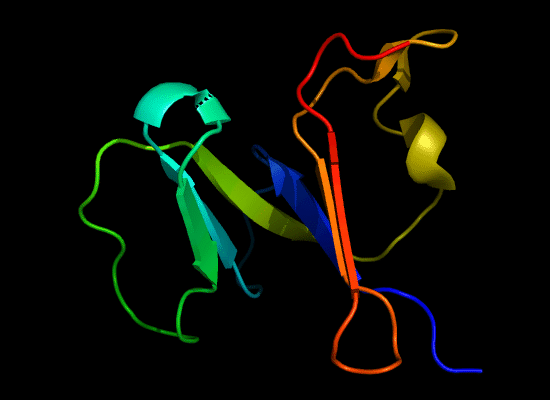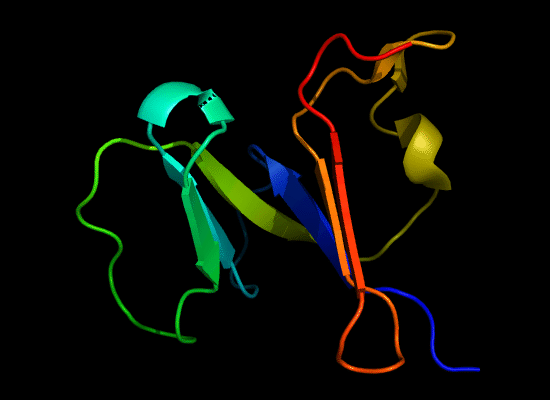
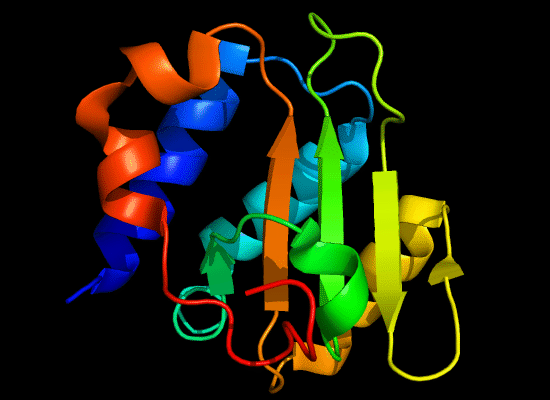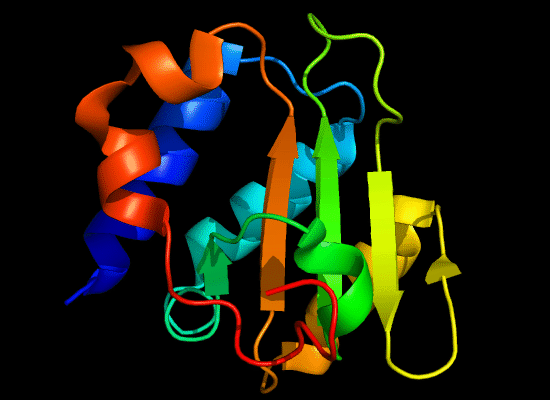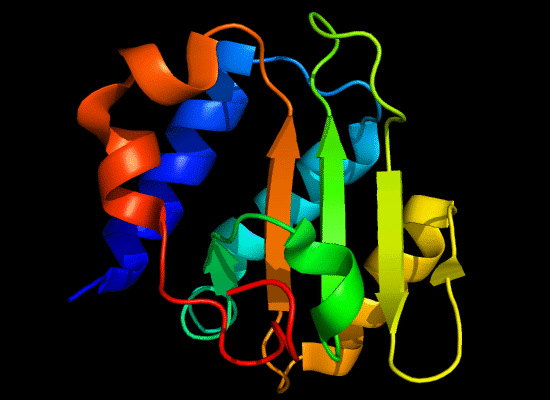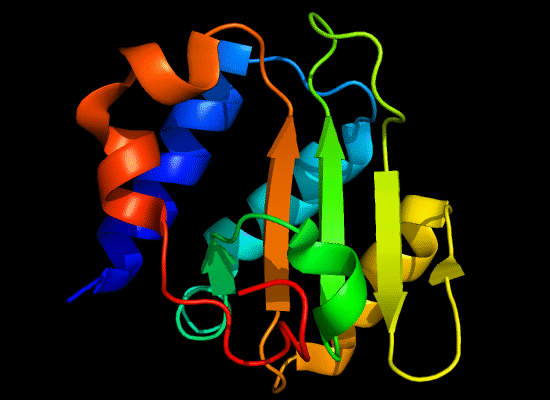
N-domain of T0397: 3d4r chain A residues -7-82
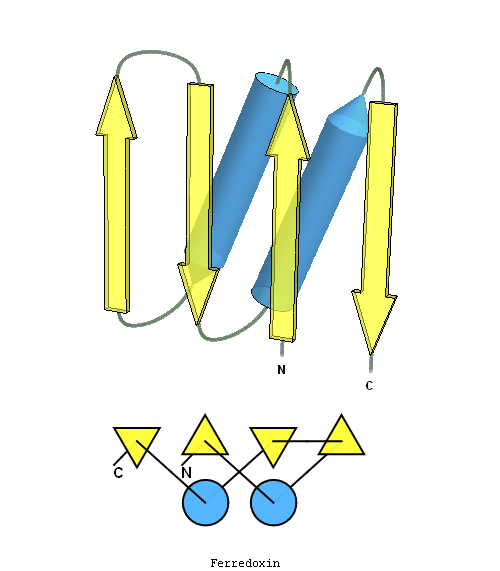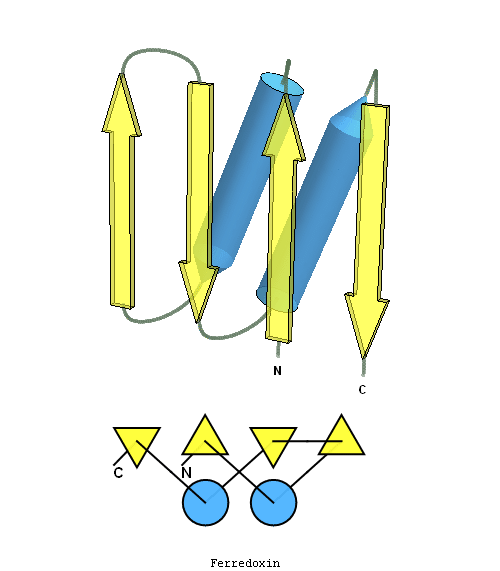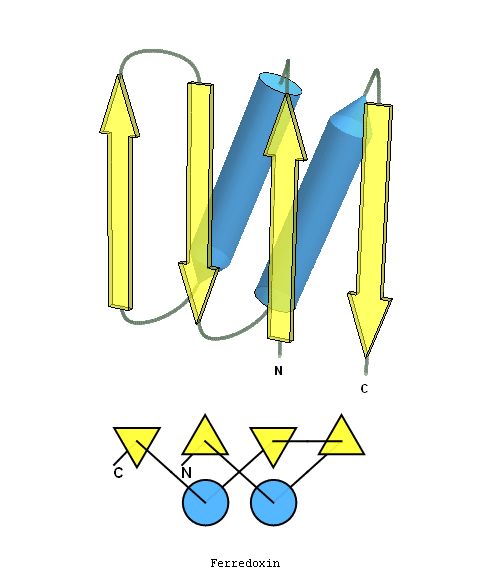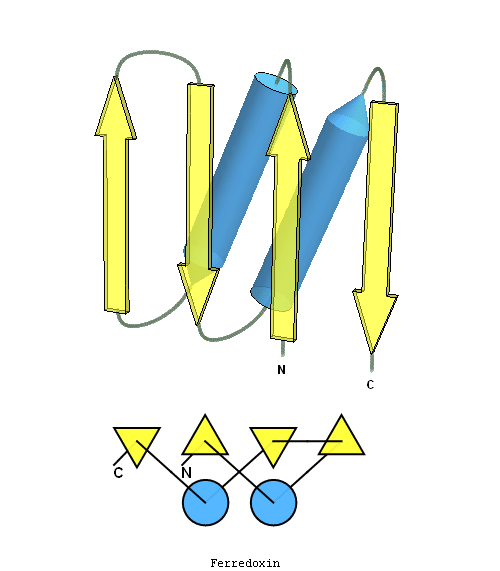
structure and topology diagrams of
ferredoxin fold – fold closest to T0397_1
N-domain of T0496:
3do9 chain A, residues 4-126
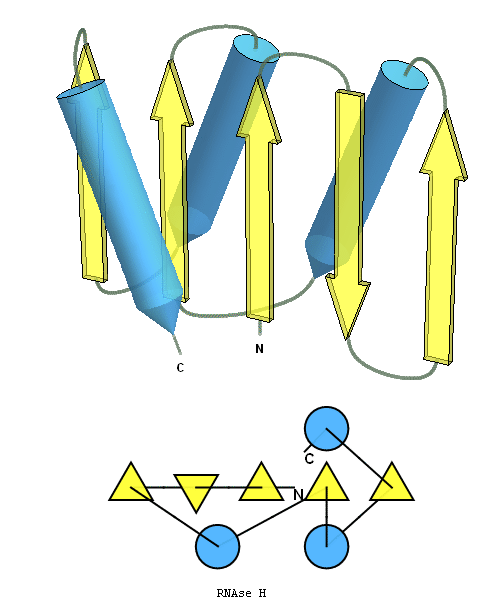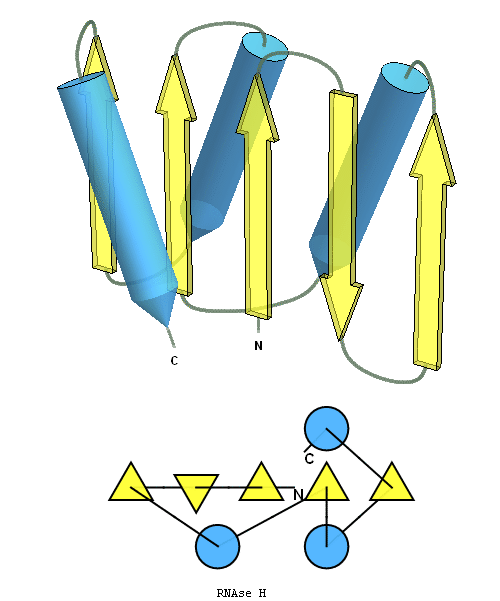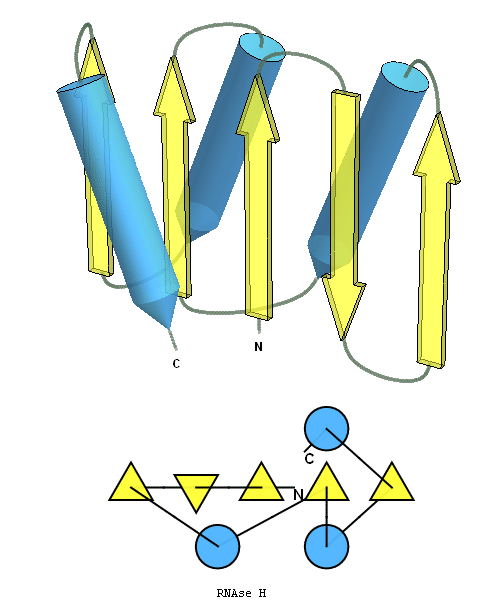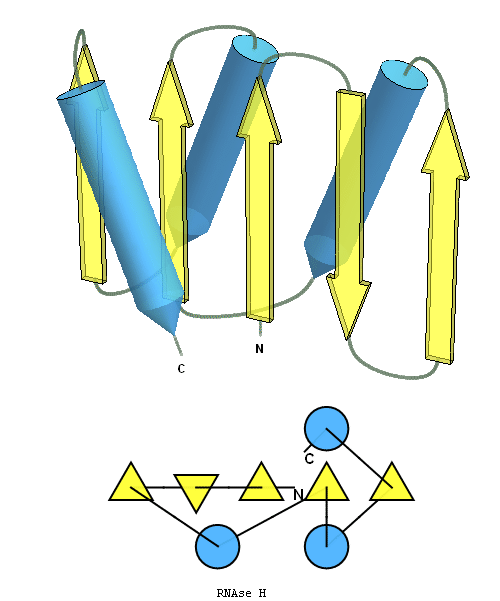
structure and topology diagrams of
RNAseH fold – fold closest to T0496_1
Nevertheless, there are some similarities between each of these domains and PDB structures. N-domain of T0397 has some topological resemblance to Ferredoxin-like fold with a curved β-sheet and α-helices deteriorated into loops. The ferredoxin-like core is elaborated with a loop and a β-strand (green) inserted into its β-hairpin and a β-strand (red) at its C-terminus. N-domain of T0496 shares similarity with RNAse H fold, and may even be viewed as a circular permutation of it.
Server predictions for both of these new folds were quite poor. However, our analysis shows that other two targets, clearly with known folds, i.e. T0407_2: an IG-like domain; and T0465: FYSH domain, were predicted very poorly as well. Apparently, as far as structure prediction is concerned, there is little difference between new folds and known folds for which templates are not detectable from sequence. Due to this observation and small number of new folds, CASP category definition should be based on a different criterion, e.g. quality of a few best server predictions.